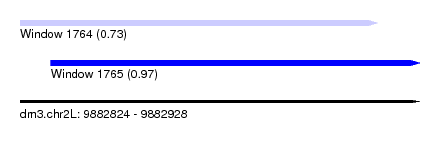
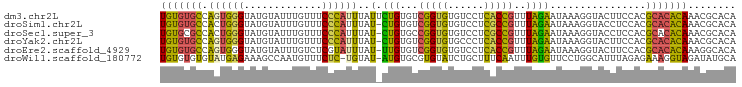
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 9,882,824 – 9,882,928 |
| Length | 104 |
| Max. P | 0.971992 |

| Location | 9,882,824 – 9,882,917 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 79.64 |
| Shannon entropy | 0.38713 |
| G+C content | 0.45140 |
| Mean single sequence MFE | -23.96 |
| Consensus MFE | -16.88 |
| Energy contribution | -15.97 |
| Covariance contribution | -0.91 |
| Combinations/Pair | 1.54 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.728284 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
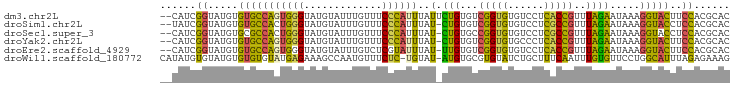
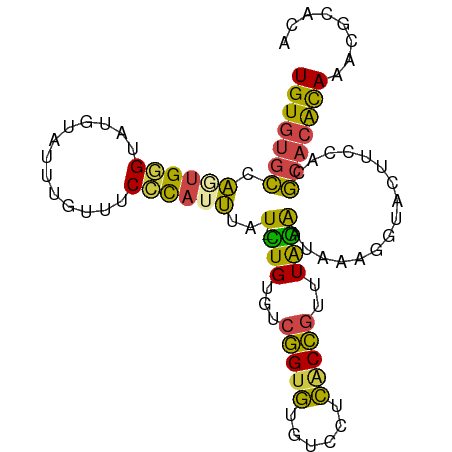
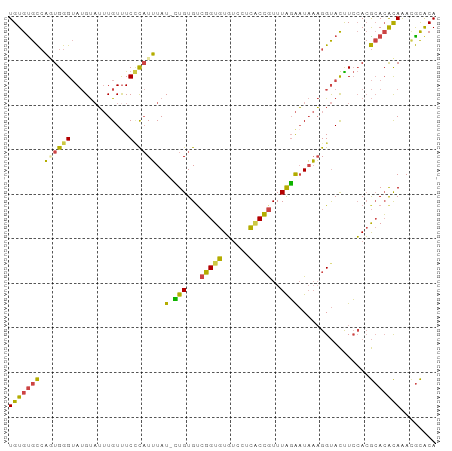
>dm3.chr2L 9882824 93 + 23011544 --CAUCGGUAUGUGUGCCAGUGGGUAUGUAUUUGUUUCCCAUUUAUUCUGUGUCGGUGUGUCCUCACCGUUUAGAAUAAAGGUACUUCCACGCAC --.....(..((.(((((..((((.............))))(((((((((...(((((......)))))..))))))))))))))...))..).. ( -28.82, z-score = -3.08, R) >droSim1.chr2L 9667183 92 + 22036055 --UAUCGGUAUGUGUGCCACUGGGUAUGUAUUUGUUUCCCAUUUAU-CUGUGUCGGUGUGUCCUCGCCGUUUAGAAUAAAGGUACCUCCACGCAC --....((...(.(((((..((((.............))))(((((-(((...(((((......)))))..))).))))))))))).))...... ( -24.02, z-score = -1.13, R) >droSec1.super_3 5328728 92 + 7220098 --CAUCGGUAUGUGCGCCACUGGGUAUGUAUUUGUUUCCCAUUUAU-CUGUGCCGGUGUGUCCUCGCCGUUUAGAAUAAAGGUACCUCCACGCAC --.........(((((.....((((((...(((((((..(((....-..))).(((((......)))))....))))))).))))))...))))) ( -21.30, z-score = 0.38, R) >droYak2.chr2L 12572550 92 + 22324452 --CAUCGGUAUGUGUGCCAGUGGGUAUGUAUUUGUUUCCCAUUUAU-CUGUGUCGGUGUGCCCUCACCGUUUAGAAUAAAGGUACUUCCACGCAC --.....(..((.(((((((((((.............))))))..(-(((...(((((......)))))..)))).....)))))...))..).. ( -25.32, z-score = -1.46, R) >droEre2.scaffold_4929 10496522 92 + 26641161 --CAUCGGUAUGUGUGCCAGUGGGUAUGUAUUUGUCUCGUAUUUAU-UUGUGUCGGUGUGUCCUCACCGUUUAGAAUAAAGGUACUUCCACGCAC --....((((....)))).(((((...(((((.........(((((-((....(((((......)))))....)))))))))))).))))).... ( -23.70, z-score = -1.30, R) >droWil1.scaffold_180772 8559463 93 + 8906247 CAUAUGUGUAUGUGUGUGUAUGAGAAAGCCAAUGUUUCUC-UGUAU-AUGUGCGUGUAUCUGCUUUCAAUUUGUGUUCCUGGCAUUUAGAGAAAG .....(((((((..(((((((((((((.......))))))-.))))-)))..)))))))...(((((..(..((((.....))))..)..))))) ( -20.60, z-score = -1.49, R) >consensus __CAUCGGUAUGUGUGCCAGUGGGUAUGUAUUUGUUUCCCAUUUAU_CUGUGUCGGUGUGUCCUCACCGUUUAGAAUAAAGGUACUUCCACGCAC ......((.....(((((..((((.............))))(((((.(((...(((((......)))))..))).))))))))))..))...... (-16.88 = -15.97 + -0.91)
| Location | 9,882,832 – 9,882,928 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 79.37 |
| Shannon entropy | 0.40011 |
| G+C content | 0.45076 |
| Mean single sequence MFE | -29.07 |
| Consensus MFE | -17.87 |
| Energy contribution | -17.10 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.971992 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
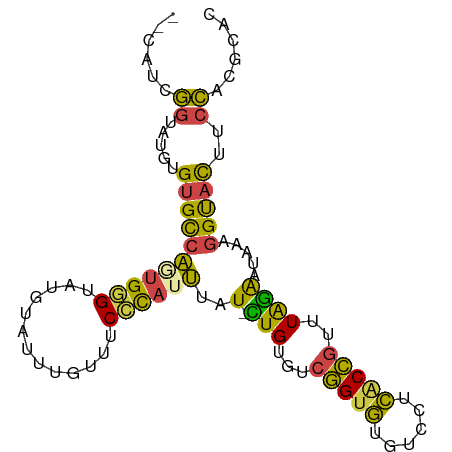
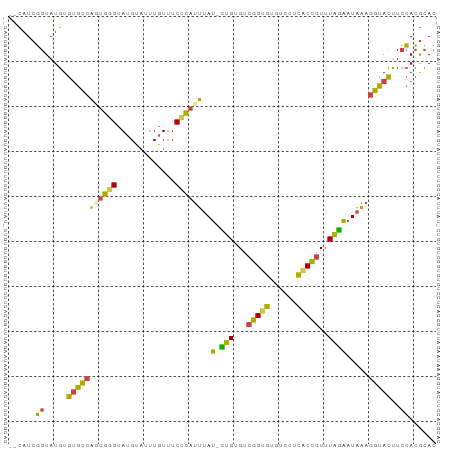
>dm3.chr2L 9882832 96 + 23011544 UGUGUGCCAGUGGGUAUGUAUUUGUUUCCCAUUUAUUCUGUGUCGGUGUGUCCUCACCGUUUAGAAUAAAGGUACUUCCACGCACACAAACGCACA (((((((..(((((...(((........((.(((((((((...(((((......)))))..)))))))))))))).))))))))))))........ ( -33.70, z-score = -3.75, R) >droSim1.chr2L 9667191 95 + 22036055 UGUGUGCCACUGGGUAUGUAUUUGUUUCCCAUUUAU-CUGUGUCGGUGUGUCCUCGCCGUUUAGAAUAAAGGUACCUCCACGCACACAAACGCACA (((((((....((((((...(((((((..(((....-..))).(((((......)))))....))))))).))))))....)))))))........ ( -26.60, z-score = -1.24, R) >droSec1.super_3 5328736 95 + 7220098 UGUGCGCCACUGGGUAUGUAUUUGUUUCCCAUUUAU-CUGUGCCGGUGUGUCCUCGCCGUUUAGAAUAAAGGUACCUCCACGCACACAAACGCACA .(((((....((((.............)))).....-.((((((((((......)))))...........((.....))..)))))....))))). ( -25.82, z-score = -0.72, R) >droYak2.chr2L 12572558 95 + 22324452 UGUGUGCCAGUGGGUAUGUAUUUGUUUCCCAUUUAU-CUGUGUCGGUGUGCCCUCACCGUUUAGAAUAAAGGUACUUCCACGCACACAAACGCACA (((((((..(((((...((((((..(((.(((....-..))).(((((......)))))....)))...)))))).))))))))))))........ ( -29.10, z-score = -1.96, R) >droEre2.scaffold_4929 10496530 95 + 26641161 UGUGUGCCAGUGGGUAUGUAUUUGUCUCGUAUUUAU-UUGUGUCGGUGUGUCCUCACCGUUUAGAAUAAAGGUACUUCCACGCACACAAAGGCACA (((((((..(((((...(((((.........(((((-((....(((((......)))))....)))))))))))).))))))))))))........ ( -29.60, z-score = -2.26, R) >droWil1.scaffold_180772 8559473 94 + 8906247 UGUGUGUGUAUGAGAAAGCCAAUGUUUCUC-UGUAU-AUGUGCGUGUAUCUGCUUUCAAUUUGUGUUCCUGGCAUUUAGAGAAAGGUAGAUAUGCA .(..(((((((((((((.......))))))-.))))-)))..)((((((((((((((..(..((((.....))))..)..)))).)))))))))). ( -29.60, z-score = -3.94, R) >consensus UGUGUGCCAGUGGGUAUGUAUUUGUUUCCCAUUUAU_CUGUGUCGGUGUGUCCUCACCGUUUAGAAUAAAGGUACUUCCACGCACACAAACGCACA (((((((...((((.............))))(((((.(((...(((((......)))))..))).)))))...........)))))))........ (-17.87 = -17.10 + -0.77)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:29:11 2011