
| Sequence ID | dm3.chrX |
|---|---|
| Location | 3,546,456 – 3,546,555 |
| Length | 99 |
| Max. P | 0.882603 |

| Location | 3,546,456 – 3,546,555 |
|---|---|
| Length | 99 |
| Sequences | 7 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 73.09 |
| Shannon entropy | 0.50224 |
| G+C content | 0.43687 |
| Mean single sequence MFE | -27.62 |
| Consensus MFE | -12.66 |
| Energy contribution | -11.98 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.53 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.882603 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
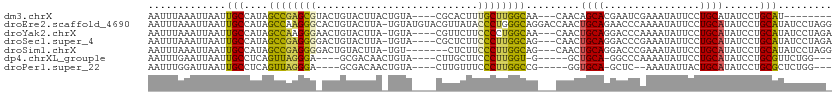
>dm3.chrX 3546456 99 + 22422827 AAUUUAAAUUAAUUGCCAUAGCCGAGCGUACUGUACUUACUGUA----CGCACUUUGCUUGGCAA---CAACAGCACGAAUCGAAAUAUUCCUGCAUAUCCUGCAU-------- ............((((((.(((.((((((((.((....)).)))----)))..)).)))))))))---.....(((.((((......)))).)))...........-------- ( -25.10, z-score = -2.86, R) >droEre2.scaffold_4690 939239 113 + 18748788 AAUUUAAAUUAAUUGCCAUAGCCAAGGGCACUGUACUUA-UGUAUGUACGUUAUACCCUGGGCAGGACCAACUGCAGAACCCAAAAUAUUCCUGCAUAUCCUGCAUAUCCUAGG ...............((.((((((.(((...(((((...-.....))))).....))))))((((((.....(((((..............)))))..)))))).....))))) ( -26.84, z-score = -0.72, R) >droYak2.chrX 16909808 106 - 21770863 AAUUUAAAUUAAUUGCCAUAGCCAAGGGAACUGUACUUA-UGUA----CGUUCUUCCCCUGGCAA---CAACUGCAGGACCCAAAAUAUUCCUGCAUAUCCUGCAUAUCCUAGA .............(((....((((.(((((.(((((...-.)))----))...))))).))))..---....(((((((..........)))))))......)))......... ( -30.20, z-score = -4.06, R) >droSec1.super_4 3029069 106 - 6179234 AAUUUAAAUUAAUUGCCAUAGCCGAGGGGACUGUACUUA-UGUA----CGCUCUUCCCUUGGCAG---CAACUGCAGGACCCGAAAUAUUCCUGCAUAUCCUGCAUAUCCUAGA ....................((((((((((.(((((...-.)))----))...)))))))))).(---((..(((((((..........))))))).....))).......... ( -33.60, z-score = -3.53, R) >droSim1.chrX 2587271 103 + 17042790 AAUUUAAAUUAAUUGCCAUAGCCGAGGGGACUGUACUUA-UGU-------CUCUUCCCUUGGCAG---CAACUGCAGGACCCGAAAUAUUCCUGCAUAUCCUGCAUAUCCUAGG ...............((...((((((((((..(.((...-.))-------)..)))))))))).(---((..(((((((..........))))))).....)))........)) ( -31.30, z-score = -2.78, R) >dp4.chrXL_group1e 8675285 96 + 12523060 AAUUUGAAUUAAUUGCCUCAGUUAGGGA----GCGACAACUGUA----CUUGCUUCCCUUGGU-G-----GCUGCA-GGCCCAAAAUAUUCCUGCAUAUCCUGCGUUCUGG--- .....((((.....(((.(....(((((----((((........----.)))).)))))..).-)-----))((((-((...........))))))........))))...--- ( -23.90, z-score = -0.19, R) >droPer1.super_22 98708 95 - 1688296 AAUUUGGAUUAAUUGCCUCAGUUAGGGA----GCGACAACUGUA----CUUGUUUCCCUUGGCCG-----GGUGCA-GCUC--AAAUAUUACUGCAUAUCCUGCGCUCUGG--- .....((........)).(((..(((((----..(((((.....----.)))))))))).(((((-----((((((-(...--........)))))...)))).)))))).--- ( -22.40, z-score = 0.50, R) >consensus AAUUUAAAUUAAUUGCCAUAGCCAAGGG_ACUGUACUUA_UGUA____CGUUCUUCCCUUGGCAG___CAACUGCAGGACCCAAAAUAUUCCUGCAUAUCCUGCAUAUCCUAG_ .............(((....((((((((...........................)))))))).........((((................))))......)))......... (-12.66 = -11.98 + -0.69)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:11:52 2011