| Sequence ID | dm3.chr2L |
|---|---|
| Location | 9,879,734 – 9,879,840 |
| Length | 106 |
| Max. P | 0.964987 |

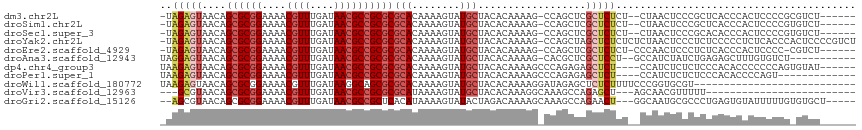
| Location | 9,879,734 – 9,879,840 |
|---|---|
| Length | 106 |
| Sequences | 11 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 73.26 |
| Shannon entropy | 0.57001 |
| G+C content | 0.51327 |
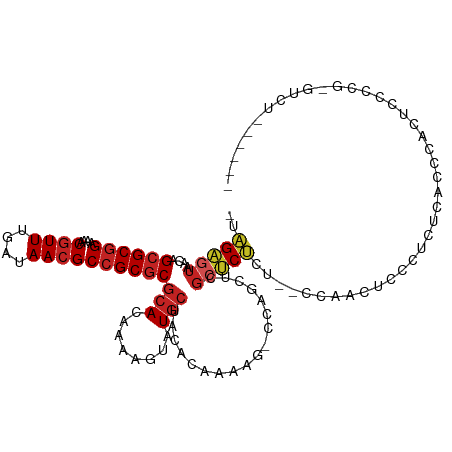
| Mean single sequence MFE | -24.32 |
| Consensus MFE | -16.21 |
| Energy contribution | -16.83 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.964987 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 9879734 106 - 23011544 -UAGAGUAACAGCGCGGAAAACGUUUGAUAACGCCGCGCGCACAAAAGUAUGCUACACAAAAG-CCAGCUCGCUCUCU--CUAACUCCCGCUCACCCACUCCCCGCGUCU------ -(((((.....((((((....((((....)))))))))).......(((..(((.........-..)))..)))..))--))).....(((.............)))...------ ( -22.02, z-score = -0.24, R) >droSim1.chr2L 9664152 106 - 22036055 -UAGAGUAACAGCGCGGAAAACGUUUGAUAACGCCGCGCGCACAAAAGUAUGCUACACAAAAG-CCAGCUCGCUCUCU--CUAACUCCCGCUCACCCACUCCCCGUGUCU------ -..((((....((((((....((((....)))))))))).......(((..(((.........-..)))..)))....--.........))))...(((.....)))...------ ( -22.20, z-score = -0.42, R) >droSec1.super_3 5325704 106 - 7220098 -UAGAGUAACAGCGCGGAAAACGUUUGAUAACGCCGCGCGCACAAAAGUAUGCUACACAAAAG-CCAGCUCGCUCUCU--CUAACUCCCGCACACCCACUCCCCGUGUCU------ -(((((.....((((((....((((....)))))))))).......(((..(((.........-..)))..)))..))--)))......((((...........))))..------ ( -22.30, z-score = -0.45, R) >droYak2.chr2L 12569484 114 - 22324452 -UAGAGUAACAGCGCGGAAAACGUUUGAUAACGCCGCGCGCACAAAAGUAUGCUACACAAAAG-CCAGCUAGCUCUCUCUCUAACUCCCUCUCCCCCUCUCACCCACUCCCCGUCU -.(((((....((((((....((((....)))))))))).......(((..(((.......))-)..))).)))))........................................ ( -21.20, z-score = -1.40, R) >droEre2.scaffold_4929 10493480 106 - 26641161 -UAGAGUAACAGCGCGGAAAACGUUUGAUAACGCCGCGCGCACAAAAGUAUGCUACACAAAAG-CCAGCUCGCUCUCU-CCCAACUCCCUCUCACCCACUCCCC-CGUCU------ -.(((((....((((((....((((....)))))))))).......(((..(((.......))-)..))).)))))..-.........................-.....------ ( -20.60, z-score = -1.26, R) >droAna3.scaffold_12943 1325315 102 - 5039921 UAGGAGUAACAGCGCGGAAAACGUUUGAUAACGCCGCGCGCACAAAAGUAUGCUACACAAAAG-CACGCUCGCUCCU--GCCAUCUAUCUGAGAGCUUUGUGUCU----------- .((((((....((((((....((((....)))))))))).......((..((((.......))-))..)).))))))--((..((.....))..)).........----------- ( -30.80, z-score = -1.12, R) >dp4.chr4_group3 11402779 106 + 11692001 UAAGAGUAACAGCGCGGAAAACGUUUGAUAACGCCGCGCGCACAAAAGUAUGCUACACAAAAGCCCAGAGAGCUCU----CCAUCUCUCUCCCACACCCCCCCAGUGUAU------ ...........((((((....((((....))))))))))((((........(((.......)))..((((((....----....))))))..............))))..------ ( -25.30, z-score = -2.71, R) >droPer1.super_1 8519301 99 + 10282868 UAAGAGUAACAGCGCGGAAAACGUUUGAUAACGCCGCGCGCACAAAAGUAUGCUACACAAAAGCCCAGAGAGCUCU----CCAUCUCUCUCCCACACCCCAGU------------- .....(((...((((((....((((....))))))))))(((........))))))..........((((((....----....)))))).............------------- ( -22.50, z-score = -2.04, R) >droWil1.scaffold_180772 8555444 89 - 8906247 UAAGAGUAACAGCGCGGAAAACGUUUGAUAAGGCAGCGCGCACAAAAGUAUGCUACACAAAAGGAUAGAGCUCUCUUUUCCCGGUGCGU--------------------------- ...........(((((.....)((((....)))).))))((((...((....))........(((.(((....)))..)))..))))..--------------------------- ( -18.70, z-score = 0.91, R) >droVir3.scaffold_12963 18505169 85 + 20206255 ---GCGUAACAGCGCGGAAAACGUUUGAUAACGCCGCGCGCAUAAAAGUAUGCUACACAAAGGCAAAGCCAGAGCU---AGCAACGUUUUU------------------------- ---(((((...((((((....((((....))))))))))(((((....)))))))).....(((...)))......---.)).........------------------------- ( -24.60, z-score = -0.90, R) >droGri2.scaffold_15126 6811865 106 - 8399593 --AGCGUAACAGCGCGGAAAACGUUUGAUAACGCCGCUCACAUAAAAGUAUACUAGACAAAAGCAAAGCCAGAACU---GGCAAUGCGCCCUGAGUGUAUUUUUGUGUGCU----- --.((......))((((....((((....)))))))).((((((((((((((((........(((..((((....)---)))..)))......))))))))))))))))..----- ( -37.34, z-score = -3.66, R) >consensus _UAGAGUAACAGCGCGGAAAACGUUUGAUAACGCCGCGCGCACAAAAGUAUGCUACACAAAAG_CCAGCUCGCUCUCU__CCAACUCCCUCUCACCCACUCCCCG_GUCU______ ..(((((....((((((....((((....))))))))))(((........)))..................)))))........................................ (-16.21 = -16.83 + 0.61)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:29:09 2011