
| Sequence ID | dm3.chrX |
|---|---|
| Location | 3,538,122 – 3,538,214 |
| Length | 92 |
| Max. P | 0.979310 |

| Location | 3,538,122 – 3,538,214 |
|---|---|
| Length | 92 |
| Sequences | 9 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 67.97 |
| Shannon entropy | 0.63913 |
| G+C content | 0.41517 |
| Mean single sequence MFE | -21.89 |
| Consensus MFE | -9.78 |
| Energy contribution | -9.44 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.02 |
| SVM RNA-class probability | 0.979310 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
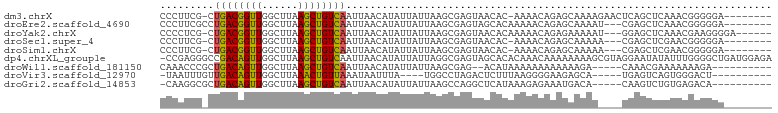
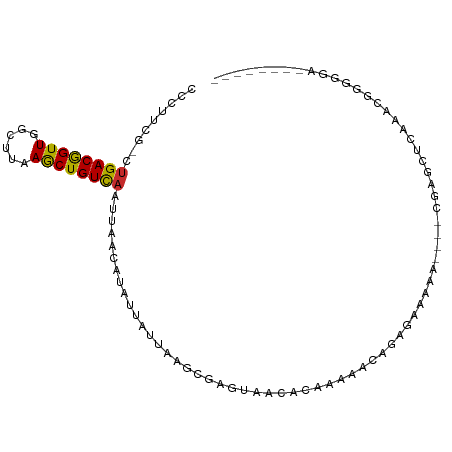
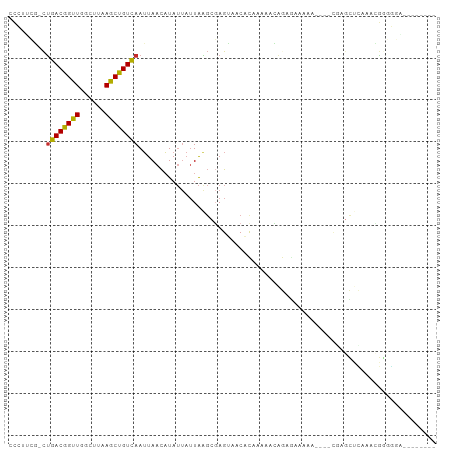
>dm3.chrX 3538122 92 - 22422827 CCCUUCG-CUGACGGUUGGCUUAAGCUGUCAAUUAACAUAUUAUUAAGCGAGUAACAC-AAAACAGAGCAAAAGAACUCAGCUCAAACGGGGGA-------- (((((((-(((((((((......)))))))(((......)))....))))).......-......((((...........))))....))))..-------- ( -23.50, z-score = -2.06, R) >droEre2.scaffold_4690 931199 91 - 18748788 CCCUUCGCCUGACGGUUGGCUUAAGCUGUCAAUUAACAUAUUAUUAAGCGAGUAGCACAAAAACAGAGCAAAAU---CGAGCUCAAACGGGGGA-------- .((((((..((((((((......))))))))................((.....)).........((((.....---...))))...)))))).-------- ( -24.40, z-score = -1.75, R) >droYak2.chrX 16900994 92 + 21770863 CCCCUCG-CUGACGGUUGGCUUAAGCUGUCAAUUAACAUAUUAUUAAGCGAGUAACACAAAAACAGAGAAAAAU---GGAGCUCAAACGAAGGGGA------ (((((((-.((((((((......))))))))..................((((..((................)---)..))))...)).))))).------ ( -24.19, z-score = -2.45, R) >droSec1.super_4 3021286 89 + 6179234 CCCUUCG-CUGACGGUUGGCUUAAGCUGUCAAUUAACAUAUUAUUAAGCGAGUAACAC-AAAACAGAGCAAAAA---CGAGCUCGAACGGGGGA-------- (((((((-(((((((((......)))))))(((......)))....))))).......-......((((.....---...))))....))))..-------- ( -24.50, z-score = -2.35, R) >droSim1.chrX 2579078 89 - 17042790 CCCUUCG-CUGACGGUUGGCUUAAGCUGUCAAUUAACAUAUUAUUAAGCGAGUAACAC-AAAACAGAGCAAAAA---CGAGCUCGAACGGGGGA-------- (((((((-(((((((((......)))))))(((......)))....))))).......-......((((.....---...))))....))))..-------- ( -24.50, z-score = -2.35, R) >dp4.chrXL_group1e 3840611 101 + 12523060 -CCGAGGGCCGACAGUUGGCUUAAGCUGUCAAUUAACAUAUUAUUAGGCGAGUAGCACACAAACAAAAAAAAGCGUAGGAAUAUAUUUGGGGCUGAUGGAGA -(((..(((((((((((......))))))).......(((((.(((.((.......................)).))).)))))......))))..)))... ( -23.80, z-score = -1.48, R) >droWil1.scaffold_181150 1142759 85 + 4952429 CAAACCCGCUGACAGUUGGCUUAAGCUGUCAAUUAACAUAUUAUUAAGCGAG--ACAUAAAAAAAAAAAAGA-----CAAACGAAAAAAAGA---------- ......(((((((((((......)))))))(((......)))....))))..--..................-----...............---------- ( -12.70, z-score = -2.17, R) >droVir3.scaffold_12970 6763261 82 - 11907090 -UAAUUUGUUGACAGUUGGCUUAAACUGUUAAAUAAUUUA----UGGCCUAGACUCUUUAAGGGGAAGAGCA-----UGAGUCAGUGGGACU---------- -(((.((((((((((((......))))))).))))).)))----((((.((..((((((.....))))))..-----)).))))........---------- ( -19.10, z-score = -1.03, R) >droGri2.scaffold_14853 4383634 86 + 10151454 -CAAGGCGCUGACAGUUGGCUUAAGCUGUCAAUUAACAUAUUAUUAAGCCAGGCUCAUAAAGAGAAAUGACA-----CAAGUCUGUGAGACA---------- -...(((..((((((((......)))))))).((((.......))))))).(.((((((...(....)(((.-----...))))))))).).---------- ( -20.30, z-score = -1.04, R) >consensus CCCUUCG_CUGACGGUUGGCUUAAGCUGUCAAUUAACAUAUUAUUAAGCGAGUAACACAAAAACAGAGAAAAA____CGAGCUCAAACGGGGGA________ .........((((((((......))))))))....................................................................... ( -9.78 = -9.44 + -0.33)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:11:50 2011