| Sequence ID | dm3.chrX |
|---|---|
| Location | 3,531,496 – 3,531,612 |
| Length | 116 |
| Max. P | 0.982011 |

| Location | 3,531,496 – 3,531,612 |
|---|---|
| Length | 116 |
| Sequences | 7 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.78 |
| Shannon entropy | 0.46690 |
| G+C content | 0.54477 |
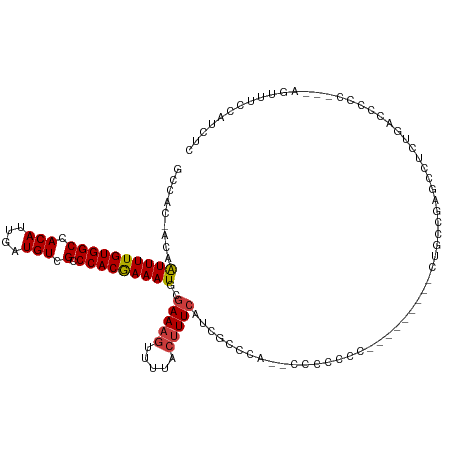
| Mean single sequence MFE | -19.09 |
| Consensus MFE | -14.42 |
| Energy contribution | -14.53 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.09 |
| SVM RNA-class probability | 0.982011 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 3531496 116 - 22422827 GCCAC-ACAAUUUUGUGGCCACAUUGAUGUCGCCCACGAAAUGCGAAAGUUUUACUUUCAUAGCCCAUCACCCCCCUCCCCCCCACUGCCGAGCCUCUGACCCCU---CGUUUCCAUCUC (((((-(......)))))).....((((((((....))).....(((((.....)))))......)))))...................((((..........))---)).......... ( -20.30, z-score = -2.33, R) >droSim1.chrX 2572698 114 - 17042790 GCCAC-ACAAUUUUGUGGCCACAUUGAUGUCGCCCACAAAAUGCGAAAGUUUUACUUUCAUAGCCCG--ACCCCCCACACCCCCACUGCCGAGCCUCUGACCCCC---AGUUUCCAUCUC ((...-...((((((((((.(((....))).).)))))))))..(((((.....)))))...))...--.....................((....(((.....)---))......)).. ( -18.30, z-score = -1.49, R) >droSec1.super_4 3014893 114 + 6179234 GCCAC-ACAAUUUUGUGGCCACAUUGAUGUCGCCCACAAAAUGCGAAAGUUUUACUUUCAUAGCCCG--ACCCCCCAUACCCCCACUGCCGAGCCUCUGACCCCC---AGCUUCCAUCUC .....-...((((((((((.(((....))).).)))))))))(((((((.....)))))...((.((--....................)).))...........---.))......... ( -18.75, z-score = -1.74, R) >droYak2.chrX 16894018 108 + 21770863 GCCAC-ACAAUUUUGUGGCCACAUUGAUGUCGCCCACGAAAUGCGAAAGUUUUACUUUCAUCGCCCA--CUUUCCC---------CAGCCGAGCCUCUGACCCCCCCAGAUUGCCAUCUC (((((-(......))))))......((((((((....((((.(((((((.....)))...))))...--.))))..---------..).)))((.((((.......))))..)))))).. ( -21.50, z-score = -2.10, R) >droEre2.scaffold_4690 924789 105 - 18748788 GCCAC-ACAAUUUUGUGGCCACAUUGAUGUCGCCCACGAAAUGCGAAAGUUUUACUUUCAUCACCCA--CUUUCCC---------CUGCCGACCCUGUGACCCCC---AGUUUCCAUCUC ....(-(((((((((((((.(((....))).).)))))))))..(((((.....)))))........--.......---------..........))))......---............ ( -19.60, z-score = -2.05, R) >droAna3.scaffold_13248 3879120 96 - 4840945 GCCAC-ACAAUUUUGUGGCCACAUUGAUGUCGCCCACGAAAUGCGAAAGUUUUACUUUCAUCGCCUU-GCCCCACU---------CCCCUGAGCCCUUCCCCACUUC------------- ((...-...((((((((((.(((....))).).)))))))))..(((((.....)))))...))...-......((---------(....)))..............------------- ( -17.10, z-score = -1.79, R) >dp4.chrXL_group1e 3831920 93 + 12523060 GCCACAACAGUUUUGUGGCCACAUUGAUGUCGCCCACGAAAUGGAAAAGUUUUGGCUUGUCCCCC----CCCCCCC---------CCCCCAACCCAGACCACGCUC-------------- ((((.(((.((((((((((.(((....))).).)))))))))......))).)))).........----.......---------.....................-------------- ( -18.10, z-score = -0.51, R) >consensus GCCAC_ACAAUUUUGUGGCCACAUUGAUGUCGCCCACGAAAUGCGAAAGUUUUACUUUCAUCGCCCA__CCCCCCC_________CUGCCGAGCCUCUGACCCCC___AGUUUCCAUCUC .........((((((((((.(((....))).).)))))))))..(((((.....)))))............................................................. (-14.42 = -14.53 + 0.10)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:11:48 2011