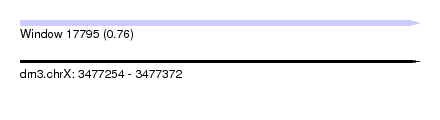
| Sequence ID | dm3.chrX |
|---|---|
| Location | 3,477,254 – 3,477,372 |
| Length | 118 |
| Max. P | 0.762335 |

| Location | 3,477,254 – 3,477,372 |
|---|---|
| Length | 118 |
| Sequences | 7 |
| Columns | 122 |
| Reading direction | forward |
| Mean pairwise identity | 79.01 |
| Shannon entropy | 0.39956 |
| G+C content | 0.38584 |
| Mean single sequence MFE | -24.24 |
| Consensus MFE | -13.90 |
| Energy contribution | -14.14 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.762335 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
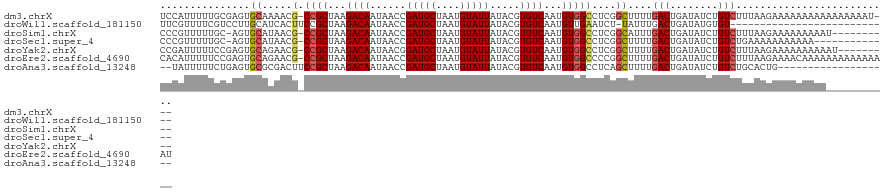
>dm3.chrX 3477254 118 + 22422827 UCCAUUUUUGCGAGUGCAAAACG-CCGCUAAGACAAUAACCGAUGCUAAUGUAUUAUACGUGUCAAUGUGGCCUCGGCUUUUGACUGAUAUCUGUCUUUAAGAAAAAAAAAAAAAAAAU--- .((..((((((....)))))).(-((((...((((......(((((....))))).....))))...)))))...))(((..(((........)))...))).................--- ( -24.80, z-score = -1.96, R) >droWil1.scaffold_181150 1069048 94 - 4952429 UUCGUUUUCGUCCUUGCAUCACUUCCGCUAAGACAAUAACCGAUGCUAAUGUAUUAUACGUGUCAAUGUUGAAUCU-UAUUUGACUGAUAUGUGU--------------------------- ...(((...(((...((.........))...)))...))).(((((....)))))((((((((((..((..(((..-.)))..))))))))))))--------------------------- ( -18.40, z-score = -1.39, R) >droSim1.chrX 2545968 110 + 17042790 CCCGUUUUUGC-AGUGCAUAACG-CCGCUAAGACAAUAACCGAUGCUAAUGUAUUAUACGUGUCAAUGUGGCCUCGGCAUUUGACUGAUAUCUGUCUUUAAGAAAAAAAAAU---------- ....((((((.-(((((.....(-((((...((((......(((((....))))).....))))...)))))....))))).(((........)))..))))))........---------- ( -23.50, z-score = -1.14, R) >droSec1.super_4 2982732 107 - 6179234 CCCGUUUUUGC-AGUGCAUAACG-CCGCUAAGACAAUAACCGAUGCUAAUGUAUUAUACGUGUCAAUGUGGCCUCGGCUUUUGACUGAUAUCUGUCUGAAAAAAAAAAA------------- ....(((((.(-((.(((....(-((((...((((......(((((....))))).....))))...))))).(((((....).))))....)))))).))))).....------------- ( -23.20, z-score = -1.30, R) >droYak2.chrX 16861136 112 - 21770863 CCGAUUUUUCCGAGUGCAGAACG-CCGCUAAGACAAUAACGGAUGCUAAUGUAUUAUACGUGUCAAUGUGGCCUCGGCUUUUGACUGAUAUCUGUCUUUAAGAAAAAAAAAAU--------- ....((((((.(((.(((((..(-((((...((((......(((((....))))).....))))...))))).(((((....).))))..))))))))...))))))......--------- ( -28.60, z-score = -2.49, R) >droEre2.scaffold_4690 891972 121 + 18748788 CACAUUUUUCCGAGUGCAGAACG-CCGCUAAGACAAUAACCGAUGCUAAUGUAUUAUACGUGUCAAUGUGGCCCCGGCUUUUGACUGAUAUCUGUCUUUAAGAAAACAAAAAAAAAAAAAAU ....(((((..(((.(((((..(-((((...((((......(((((....))))).....))))...)))))..((((....).)))...))))))))..)))))................. ( -25.10, z-score = -1.96, R) >droAna3.scaffold_13248 3820233 101 + 4840945 --UAUUUUUCUGAGUGCGCGACUUCCGCUAAGACAAUAACCGAUGCUAAUGUAUUAUACGUGUCAAUGUGGCCUCAGCUUUUGACUGAUAUCUGUCUGCACUG------------------- --..........((((((.(((..((((...((((......(((((....))))).....))))...))))..(((((....).)))).....))))))))).------------------- ( -26.10, z-score = -2.28, R) >consensus CCCAUUUUUGCGAGUGCAUAACG_CCGCUAAGACAAUAACCGAUGCUAAUGUAUUAUACGUGUCAAUGUGGCCUCGGCUUUUGACUGAUAUCUGUCUUUAAGAAAAAAAAA___________ ..............................(((((......(((((....)))))...((.(((((...(((....))).))))))).....)))))......................... (-13.90 = -14.14 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:11:44 2011