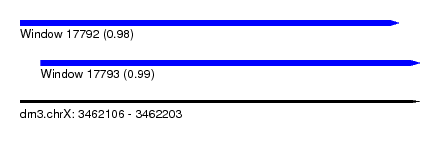
| Sequence ID | dm3.chrX |
|---|---|
| Location | 3,462,106 – 3,462,203 |
| Length | 97 |
| Max. P | 0.988125 |

| Location | 3,462,106 – 3,462,198 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 72.78 |
| Shannon entropy | 0.44110 |
| G+C content | 0.58716 |
| Mean single sequence MFE | -25.46 |
| Consensus MFE | -16.12 |
| Energy contribution | -16.56 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.979649 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
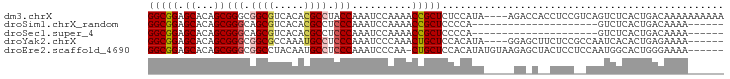
>dm3.chrX 3462106 92 + 22422827 CUAAUGGCGGAGCACAGCGGGCGGCGUCACACGCCUACCAAAUCCAAAACCGCUCUCCAUA----AGACCACCUCCGUCAGUCUCACUGACAAAAA ....((((((((......((..((((.....))))..))............(.(((.....----))).)..))))))))(((.....)))..... ( -26.20, z-score = -2.68, R) >droSim1.chrX_random 1465447 70 + 5698898 CUAAUGGCGGAGCACAGCGGGCAGCGUCACACGCCUCCCAAAUCCAAAACCGCUCCCCA---------------------GUCUCACUGAC----- ....(((.(((((.....(((..(((.....)))..)))...(....)...))))))))---------------------(((.....)))----- ( -20.10, z-score = -1.73, R) >droSec1.super_4 2967551 70 - 6179234 CUAAUGGCGGAGCACAGCGGGCAGCGUCACACGCCUCCCAAAUCCAAAACCGCUCCCCA---------------------GUCUCACUGAC----- ....(((.(((((.....(((..(((.....)))..)))...(....)...))))))))---------------------(((.....)))----- ( -20.10, z-score = -1.73, R) >droYak2.chrX 16844803 87 - 21770863 CUAAUGGCGGAGCACAGCGGGCGGCGCCAAAUGCCUCCCAAAUCCCAAACUGCUCCACAUA----GGAGCUUCUCCGCCAAUCACACUGAG----- ....((((((((......(((.((((.....)))).)))............(((((.....----)))))..))))))))...........----- ( -35.00, z-score = -4.21, R) >droEre2.scaffold_4690 874465 90 + 18748788 CUAAUGGCGGAGCACAGCGGGCGGCCUACAAUGCCUCCCAAAUCCCAA-CUGCUCCACAUAUGUAAGAGCUACUCCUCCAAUGGCACUGGG----- (((.(((.((((......(((.(((.......))).))).........-..((((.((....))..))))..)))).))).))).......----- ( -25.90, z-score = -0.32, R) >consensus CUAAUGGCGGAGCACAGCGGGCGGCGUCACACGCCUCCCAAAUCCAAAACCGCUCCCCAUA_____GA_C__CUCC__CAGUCUCACUGAC_____ .....(((((.((...))(((.((((.....)))).)))..........))))).......................................... (-16.12 = -16.56 + 0.44)

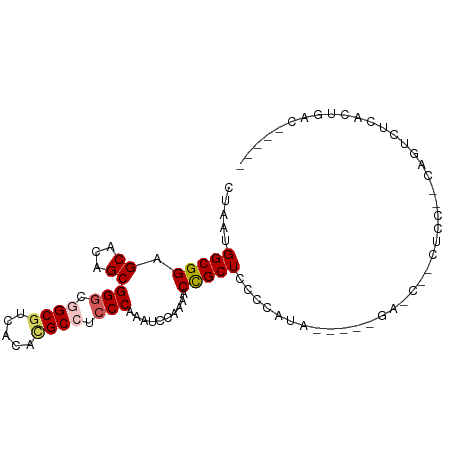
| Location | 3,462,111 – 3,462,203 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 72.14 |
| Shannon entropy | 0.44862 |
| G+C content | 0.58081 |
| Mean single sequence MFE | -25.00 |
| Consensus MFE | -15.92 |
| Energy contribution | -16.36 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.31 |
| SVM RNA-class probability | 0.988125 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 3462111 92 + 22422827 GGCGGAGCACAGCGGGCGGCGUCACACGCCUACCAAAUCCAAAACCGCUCUCCAUA----AGACCACCUCCGUCAGUCUCACUGACAAAAAAAAAA (((((((......((..((((.....))))..))............(.(((.....----))).)..))))))).(((.....))).......... ( -25.20, z-score = -2.82, R) >droSim1.chrX_random 1465452 69 + 5698898 GGCGGAGCACAGCGGGCAGCGUCACACGCCUCCCAAAUCCAAAACCGCUCCCCA---------------------GUCUCACUGACAAAA------ ((.(((((.....(((..(((.....)))..)))...(....)...))))))).---------------------(((.....)))....------ ( -20.80, z-score = -2.35, R) >droSec1.super_4 2967556 69 - 6179234 GGCGGAGCACAGCGGGCAGCGUCACACGCCUCCCAAAUCCAAAACCGCUCCCCA---------------------GUCUCACUGACAAAA------ ((.(((((.....(((..(((.....)))..)))...(....)...))))))).---------------------(((.....)))....------ ( -20.80, z-score = -2.35, R) >droYak2.chrX 16844808 86 - 21770863 GGCGGAGCACAGCGGGCGGCGCCAAAUGCCUCCCAAAUCCCAAACUGCUCCACAUA----GGAGCUUCUCCGCCAAUCACACUGAGAAAA------ (((((((......(((.((((.....)))).)))............(((((.....----)))))..)))))))................------ ( -34.00, z-score = -3.82, R) >droEre2.scaffold_4690 874470 89 + 18748788 GGCGGAGCACAGCGGGCGGCCUACAAUGCCUCCCAAAUCCCAA-CUGCUCCACAUAUGUAAGAGCUACUCCUCCAAUGGCACUGGGAAAA------ .((........))(((.(((.......))).)))...(((((.-..((((.((....))..))))....((......))...)))))...------ ( -24.20, z-score = 0.20, R) >consensus GGCGGAGCACAGCGGGCGGCGUCACACGCCUCCCAAAUCCAAAACCGCUCCCCAUA_____GA_C__CUCC__CAGUCUCACUGACAAAA______ (((((.((...))(((.((((.....)))).)))..........)))))............................................... (-15.92 = -16.36 + 0.44)
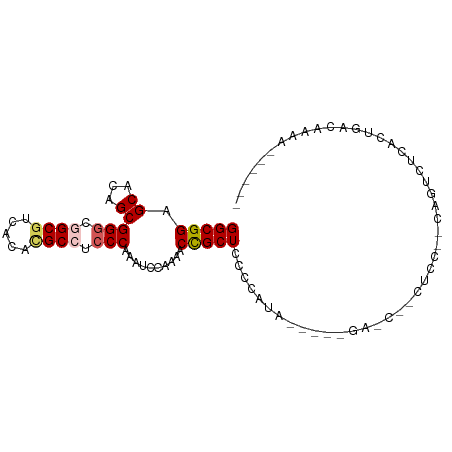
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:11:42 2011