
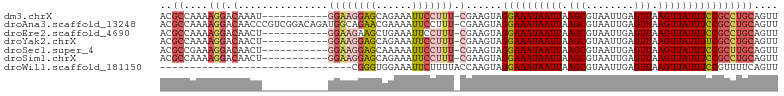
| Sequence ID | dm3.chrX |
|---|---|
| Location | 3,445,075 – 3,445,166 |
| Length | 91 |
| Max. P | 0.967089 |

| Location | 3,445,075 – 3,445,166 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 83.51 |
| Shannon entropy | 0.31502 |
| G+C content | 0.38280 |
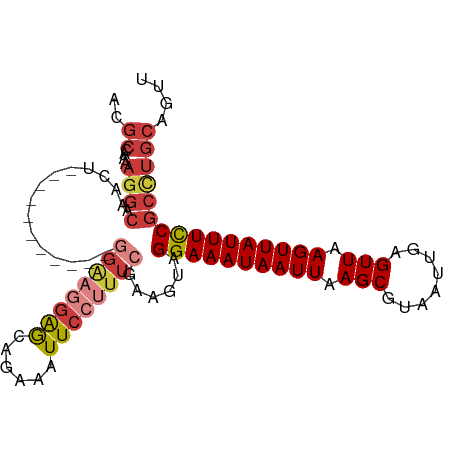
| Mean single sequence MFE | -23.61 |
| Consensus MFE | -14.60 |
| Energy contribution | -15.46 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.967089 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 3445075 91 - 22422827 ACGCCAAAAGGACAAAU-----------GGAAGGAGCAGAAAUUCCUUU-CGAAGUAGGAAAUAAUUAAGCGUAAUUGAGUUAAGUUAUUUCCGCCUGCAGUU ..((....(((.(...(-----------((((((((......)))))))-)).....((((((((((.(((........))).)))))))))))))))).... ( -25.20, z-score = -3.23, R) >droAna3.scaffold_13248 3766018 102 - 4840945 ACGCCAAAAGGACAACCCGUCGGACAGAUGGCAGAACGAAAAUUCCUUU-CGAAGUAGGAAAUAAUUAAGCGUAAUUGAGUUAAGUUAUUUCCGCCUGCAGUU ..(((....)).)....((((.....))))((((..(((((.....)))-)).....((((((((((.(((........))).))))))))))..)))).... ( -26.10, z-score = -2.08, R) >droEre2.scaffold_4690 857668 91 - 18748788 ACGCCAAAAGGACAACU-----------GGAAGAAGCUGAAAUUCCUUU-CGAAGUAGGAAAUAAUUAAGCGUAAUUGAGUUAAGUUAUUUCCGCCUGCAGUU ..(((.((((((((.((-----------......)).))....))))))-.).((..((((((((((.(((........))).))))))))))..)))).... ( -19.70, z-score = -1.07, R) >droYak2.chrX 16828186 91 + 21770863 ACGCCAAAAGGACAACU-----------GGAAGGAGCAGAAAUUCCUUU-CGAAGUAGGAAAUAAUUAAGCGUAAUUGAGUUAAGUUAUUUUCGCCUGCAGUU ..(((....)).)((((-----------((((((((......)))))))-)...(((((((((((((.(((........))).))))))))...))))))))) ( -22.40, z-score = -1.92, R) >droSec1.super_4 2950980 91 + 6179234 ACGCCGAAAGGACAACU-----------GGAAGGAGCAAAAAUUCCUUU-CGAAGUAGGAAAUAAUUAAGCGUAAUUGAGUUAAGUUAUUUCCGCUUGCAGUU ..(((....)).)((((-----------((((((((......)))))))-).((((.((((((((((.(((........))).))))))))))))))..)))) ( -28.40, z-score = -3.86, R) >droSim1.chrX 2515490 91 - 17042790 ACGCCAAAAGGACAACU-----------GGAAGGAGCAGAAAUUCCUUU-CGAAGUAGGAAAUAAUUAAGCGUAAUUGAGUUAAGUUAUUUCCGCCUGCAGUU ..((....(((.(...(-----------((((((((......)))))))-)).....((((((((((.(((........))).)))))))))))))))).... ( -25.00, z-score = -2.77, R) >droWil1.scaffold_181150 4463277 71 + 4952429 --------------------------------CGGGUGGAAAUUCUUUUACCAAGUAGGAAAUAAUUAAGCGUAAUUGAGUUAAGUUAUUUCCGUUUUCAGUU --------------------------------..(((((((....))))))).....((((((((((.(((........))).)))))))))).......... ( -18.50, z-score = -3.72, R) >consensus ACGCCAAAAGGACAACU___________GGAAGGAGCAGAAAUUCCUUU_CGAAGUAGGAAAUAAUUAAGCGUAAUUGAGUUAAGUUAUUUCCGCCUGCAGUU ........(((..................(((((((......)))))))........((((((((((.(((........))).)))))))))).)))...... (-14.60 = -15.46 + 0.86)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:11:40 2011