| Sequence ID | dm3.chrX |
|---|---|
| Location | 3,431,975 – 3,432,070 |
| Length | 95 |
| Max. P | 0.551912 |

| Location | 3,431,975 – 3,432,070 |
|---|---|
| Length | 95 |
| Sequences | 10 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 67.96 |
| Shannon entropy | 0.64819 |
| G+C content | 0.45174 |
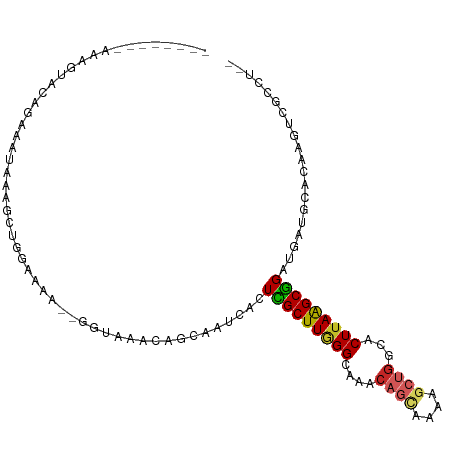
| Mean single sequence MFE | -21.70 |
| Consensus MFE | -11.75 |
| Energy contribution | -11.81 |
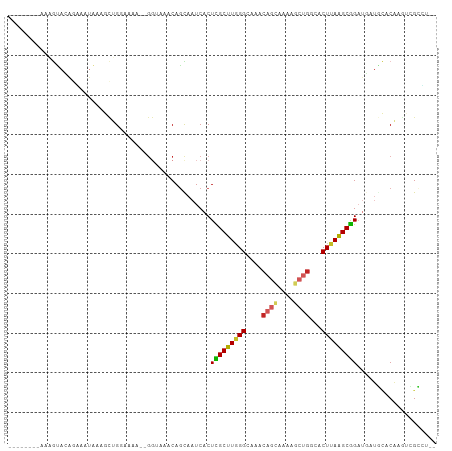
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.38 |
| Mean z-score | -0.44 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.551912 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
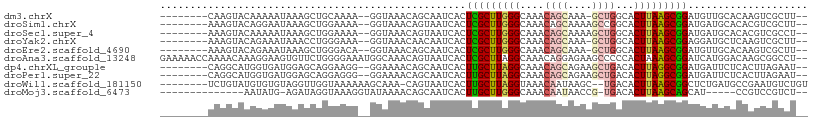
>dm3.chrX 3431975 95 + 22422827 --------CAAGUACAAAAAUAAAGCUGCAAAA--GGUAAACAGCAAUCACUCGCUUGGGCAAACAGCAAA-GCUGGCACUUAAGCGGAUGUUGCACAAGUCGCUU-- --------..............((((.((....--(.....).((((.((..((((((((....((((...-))))...))))))))..))))))....)).))))-- ( -26.00, z-score = -1.68, R) >droSim1.chrX 2500806 96 + 17042790 --------AAAGUACAGGAAUAAAGCUGGAAAA--GGUAAACAGUAAUCACUCGCUUGGGCAAACAGCAAAAGCCGGCACUUAAGCGGAUGAUGCACACGUCGCUU-- --------.((((((.........((((.....--......)))).((((..((((((((....(.((....)).)...))))))))..))))......)).))))-- ( -21.80, z-score = -0.38, R) >droSec1.super_4 2938101 96 - 6179234 --------AAAGUACAAAAAUAAAGCUGGAAAA--GGUAAACAGUAAUCACUCGCUUGGGCAAACAGCAAAAGCUGGCACUUAAGCGGAUGAUGCACACGUCGCCU-- --------........................(--((..(...((.((((..((((((((....((((....))))...))))))))..))))..))...)..)))-- ( -26.00, z-score = -2.20, R) >droYak2.chrX 16815022 95 - 21770863 --------AAAGUACAGAAAUAAACCUGGGAAA--GGUAAACAACAAUCACUCGCUUGGGCAAACAGCAAA-GCUGGCACUUAAGCGGAGGAUGCUCAAGUCGCUU-- --------.((((((........((((.....)--)))........(((..(((((((((....((((...-))))...)))))))))..)))......)).))))-- ( -23.20, z-score = -1.18, R) >droEre2.scaffold_4690 844023 95 + 18748788 --------AAAGUACAGAAAUAAAGCUGGGACA--GGUAAACAGCAAUCACUCGCUUGGGCAAACAGCAAA-GCUGGCACUUAAGCGGAUGUUGCACAAGUCGCUU-- --------.((((.(((........))).(((.--(.....).((((.((..((((((((....((((...-))))...))))))))..))))))....)))))))-- ( -27.60, z-score = -1.99, R) >droAna3.scaffold_13248 3705650 106 + 4840945 GAAAAACCAAAACAAAGGAAGUGUUCUGGGGAAAUGGCAAACAGUAAUCACUCGCUUAGGCAAACAGGAGAAGCCCCCACUAAAGCGGAUCAUGGACAAGCGGCCU-- ......(((.((((.......)))).)))......(((....(((....)))(((((.(((...........))).((.(....).)).........)))))))).-- ( -18.90, z-score = 1.67, R) >dp4.chrXL_group1e 3727622 96 - 12523060 --------CAGGCAUGGUGAUGGAGCAGGAAGG--GGAAAACAGCAAUCACUUGCUUAGGCAAACAGCAGAAGCUGACACUUAGGCGGAUGAUUCUCACUUAGAAU-- --------.......(((((.(((((.(.....--......).))..(((..((((((((....((((....))))...))))))))..)))))))))))......-- ( -21.80, z-score = -0.51, R) >droPer1.super_22 1468983 96 + 1688296 --------CAGGCAUGGUGAUGGAGCAGGAGGG--GGAAAACAGCAAUCACUUGCUUAGGCAAACAGCAGAAGCUGACACUUAGGCGGAUGAUUCUCACUUAGAAU-- --------...((..((((...(((((((.((.--.(.......)..)).))))))).......((((....)))).))))...)).....(((((.....)))))-- ( -22.10, z-score = -0.50, R) >droWil1.scaffold_181150 522182 97 + 4952429 --------UCUGUAUGUGUGUAGGUUGGUAAAAAAGCAAA-CAGUAAUCACUUGCUUAGGUAAACAAUAAGC--UGACACUUAAGCGGCUCUGAUGCCGAAUGUCUGU --------...............(((........)))..(-(((((.((....((((((((...((......--))..))))))))(((......))))).)).)))) ( -18.40, z-score = 0.89, R) >droMoj3.scaffold_6473 8945017 85 + 16943266 --------------AAUAUG-AGAUAGGUAAAGGUAUAAAACAGCAAUCACUUGCUUGGGCAAACAAUAACCG-UGACACUUAAGCAGCAU-----CCGUCCGUCU-- --------------...(((-.((..(((....((........))......(((((((((...((.......)-)....))))))))).))-----)..)))))..-- ( -11.20, z-score = 1.46, R) >consensus ________AAAGUACAGAAAUAAAGCUGGAAAA__GGUAAACAGCAAUCACUCGCUUGGGCAAACAGCAAAAGCUGGCACUUAAGCGGAUGAUGCACAAGUCGCCU__ ...................................................(((((((((....((((....))))...))))))))).................... (-11.75 = -11.81 + 0.06)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:11:39 2011