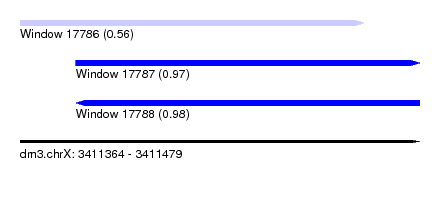
| Sequence ID | dm3.chrX |
|---|---|
| Location | 3,411,364 – 3,411,479 |
| Length | 115 |
| Max. P | 0.978287 |

| Location | 3,411,364 – 3,411,463 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 81.13 |
| Shannon entropy | 0.35569 |
| G+C content | 0.50168 |
| Mean single sequence MFE | -33.54 |
| Consensus MFE | -17.34 |
| Energy contribution | -18.21 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.564987 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 3411364 99 + 22422827 CUUGAGUUGGCCGAAGGUGACAAAAAUUCUGAACCCAGGCGUGCUGUAAGCGGUUAGUCG---GCUUACAGUGCGCUUCACGGGCAUAGGAUU-GGACCUAAA ........((((...(....)...(((((((..((((((((..((((((((.(.....).---))))))))..)))))...)))..)))))))-)).)).... ( -40.40, z-score = -3.51, R) >droSim1.chrX 2480861 99 + 17042790 CUUGAGUUGGCCGAAGGUGACAAAAAUUCUGAACCCAUGCGUGCUGUAAGCGGUUAGUCG---GCUUACAGUGCGCUUCACGGGCAUAGGAUU-GGACCUAAA ........((((...(....)...(((((((..(((..(((..((((((((.(.....).---))))))))..))).....)))..)))))))-)).)).... ( -38.40, z-score = -3.16, R) >droSec1.super_4 2918155 99 - 6179234 CUUGAGUUGGCCGAAGGUGACAAAAAUUCUGAACCCAUACGUGCUGUAAGCGGUUAGUCG---GCUUACAGUGCGCUUCACGGGCAUAGGAUU-GGACCUAAA ........((((...(....)...(((((((..(((...((..((((((((.(.....).---))))))))..))......)))..)))))))-)).)).... ( -34.10, z-score = -2.22, R) >droYak2.chrX 16794185 99 - 21770863 CUUGAGUUGGUCGAAGGUGACAAAAAUUCUGAACCCAUGCGUGCUGUAAGCGGUUAGUCG---GCUUACAGUGCGCUUCACGGGCACAGGUUU-ACACCUAAA .((((.....))))(((((....((((.(((..(((..(((..((((((((.(.....).---))))))))..))).....)))..)))))))-.)))))... ( -35.90, z-score = -2.57, R) >droEre2.scaffold_4690 819637 100 + 18748788 CUUGAGCUGGUCGAAGGUGACAAAAAUUCUGAACCCAUGCGUGUUGCAAGCGGUUAGUCG---GCUUACAGUGCGCUUCACGGGCACAGGUUUUAGACCUAAA ........((((...(....).(((((.(((..(((..(((..(((.((((.(.....).---)))).)))..))).....)))..)))))))).)))).... ( -31.60, z-score = -0.90, R) >droAna3.scaffold_13248 3670982 82 + 4840945 CUUGAGCUCUUCGAAGGCGACAAAAAGUCUGAACACACGCGUGUUGCAAGUUCCAAGUUGCAAGUUGCCAGUUCACUGCGUG--------------------- ..((((((.......((((((..........(((((....)))))((((.(....).))))..)))))))))))).......--------------------- ( -20.81, z-score = 0.48, R) >consensus CUUGAGUUGGCCGAAGGUGACAAAAAUUCUGAACCCAUGCGUGCUGUAAGCGGUUAGUCG___GCUUACAGUGCGCUUCACGGGCAUAGGAUU_GGACCUAAA .......((.(((..(((..............)))...(((..((((((((............))))))))..)))....))).))................. (-17.34 = -18.21 + 0.86)

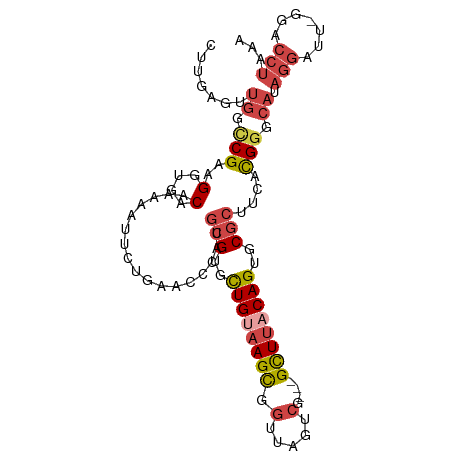
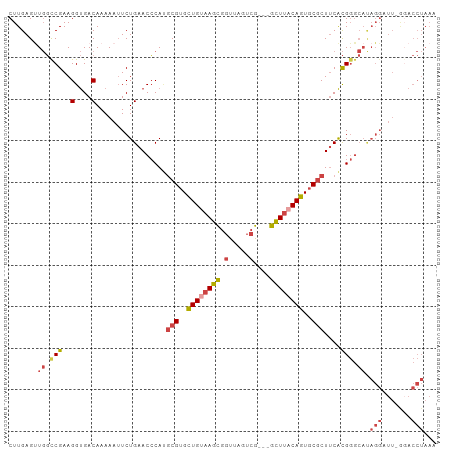
| Location | 3,411,380 – 3,411,479 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 93.40 |
| Shannon entropy | 0.11103 |
| G+C content | 0.49785 |
| Mean single sequence MFE | -33.76 |
| Consensus MFE | -31.12 |
| Energy contribution | -31.52 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.968449 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 3411380 99 + 22422827 GUGACAAAAAUUCUGAACCCAGGCGUGCUGUAAGCGGUUAGUCGGCUUACAGUGCGCUUCACGGGCAUAGGAUU-GGACCUAAACUGCCUGAGUGACAAA ((.((...............(((((..((((((((.(.....).))))))))..)))))..((((((((((...-...))))...)))))).)).))... ( -40.60, z-score = -3.98, R) >droSim1.chrX 2480877 95 + 17042790 GUGACAAAAAUUCUGAACCCAUGCGUGCUGUAAGCGGUUAGUCGGCUUACAGUGCGCUUCACGGGCAUAGGAUU-GGACCUAAACUGUCUGAGUGA---- ...((...(((((((..(((..(((..((((((((.(.....).))))))))..))).....)))..)))))))-((((.......))))..))..---- ( -34.30, z-score = -2.67, R) >droSec1.super_4 2918171 95 - 6179234 GUGACAAAAAUUCUGAACCCAUACGUGCUGUAAGCGGUUAGUCGGCUUACAGUGCGCUUCACGGGCAUAGGAUU-GGACCUAAACUGUCUGAGUGA---- ...((...(((((((..(((...((..((((((((.(.....).))))))))..))......)))..)))))))-((((.......))))..))..---- ( -30.00, z-score = -1.64, R) >droYak2.chrX 16794201 95 - 21770863 GUGACAAAAAUUCUGAACCCAUGCGUGCUGUAAGCGGUUAGUCGGCUUACAGUGCGCUUCACGGGCACAGGUUU-ACACCUAAACUGCCUGAGUGA---- ...................((((((..((((((((.(.....).))))))))..)))....((((((.((((..-..))))....)))))).))).---- ( -35.40, z-score = -2.64, R) >droEre2.scaffold_4690 819653 96 + 18748788 GUGACAAAAAUUCUGAACCCAUGCGUGUUGCAAGCGGUUAGUCGGCUUACAGUGCGCUUCACGGGCACAGGUUUUAGACCUAAACUGCCUGAGUGA---- ...................((((((..(((.((((.(.....).)))).)))..)))....((((((.(((((...)))))....)))))).))).---- ( -28.50, z-score = -0.38, R) >consensus GUGACAAAAAUUCUGAACCCAUGCGUGCUGUAAGCGGUUAGUCGGCUUACAGUGCGCUUCACGGGCAUAGGAUU_GGACCUAAACUGCCUGAGUGA____ ......................(((..((((((((.(.....).))))))))..))).(((((((((((((.......))))...)))))..)))).... (-31.12 = -31.52 + 0.40)

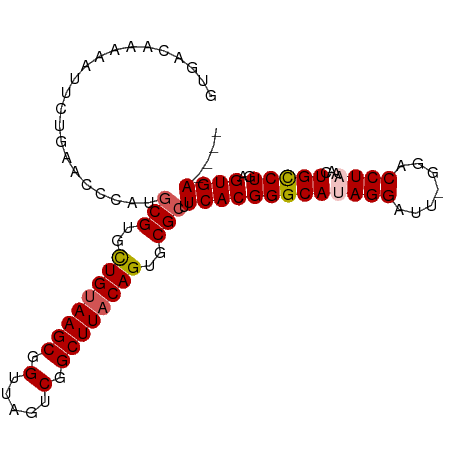

| Location | 3,411,380 – 3,411,479 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 93.40 |
| Shannon entropy | 0.11103 |
| G+C content | 0.49785 |
| Mean single sequence MFE | -29.66 |
| Consensus MFE | -28.12 |
| Energy contribution | -27.72 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.95 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.978287 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 3411380 99 - 22422827 UUUGUCACUCAGGCAGUUUAGGUCC-AAUCCUAUGCCCGUGAAGCGCACUGUAAGCCGACUAACCGCUUACAGCACGCCUGGGUUCAGAAUUUUUGUCAC ((((..((((((((.((.((((...-...)))).))..(((.......((((((((.(.....).))))))))))))))))))).))))........... ( -31.41, z-score = -2.44, R) >droSim1.chrX 2480877 95 - 17042790 ----UCACUCAGACAGUUUAGGUCC-AAUCCUAUGCCCGUGAAGCGCACUGUAAGCCGACUAACCGCUUACAGCACGCAUGGGUUCAGAAUUUUUGUCAC ----.......(((((..((((...-...)))).((((((...(((..((((((((.(.....).))))))))..))))))))).........))))).. ( -30.00, z-score = -3.15, R) >droSec1.super_4 2918171 95 + 6179234 ----UCACUCAGACAGUUUAGGUCC-AAUCCUAUGCCCGUGAAGCGCACUGUAAGCCGACUAACCGCUUACAGCACGUAUGGGUUCAGAAUUUUUGUCAC ----.......(((((..((((...-...)))).((((((...(((..((((((((.(.....).))))))))..))))))))).........))))).. ( -28.60, z-score = -2.77, R) >droYak2.chrX 16794201 95 + 21770863 ----UCACUCAGGCAGUUUAGGUGU-AAACCUGUGCCCGUGAAGCGCACUGUAAGCCGACUAACCGCUUACAGCACGCAUGGGUUCAGAAUUUUUGUCAC ----.......(((((((((....)-))))(((.((((((...(((..((((((((.(.....).))))))))..))))))))).)))......)))).. ( -31.50, z-score = -2.00, R) >droEre2.scaffold_4690 819653 96 - 18748788 ----UCACUCAGGCAGUUUAGGUCUAAAACCUGUGCCCGUGAAGCGCACUGUAAGCCGACUAACCGCUUGCAACACGCAUGGGUUCAGAAUUUUUGUCAC ----.......(((((......(((..(((((((((..(((.....)))(((((((.(.....).)))))))....))))))))).)))....))))).. ( -26.80, z-score = -0.77, R) >consensus ____UCACUCAGGCAGUUUAGGUCC_AAUCCUAUGCCCGUGAAGCGCACUGUAAGCCGACUAACCGCUUACAGCACGCAUGGGUUCAGAAUUUUUGUCAC ...........(((((..((((.......)))).((((((...(((..((((((((.(.....).))))))))..))))))))).........))))).. (-28.12 = -27.72 + -0.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:11:38 2011