
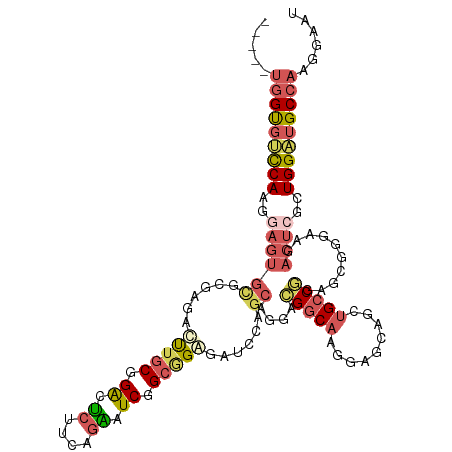
| Sequence ID | dm3.chrX |
|---|---|
| Location | 3,379,943 – 3,380,054 |
| Length | 111 |
| Max. P | 0.818749 |

| Location | 3,379,943 – 3,380,054 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 70.68 |
| Shannon entropy | 0.52517 |
| G+C content | 0.61080 |
| Mean single sequence MFE | -45.58 |
| Consensus MFE | -22.02 |
| Energy contribution | -23.82 |
| Covariance contribution | 1.80 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.818749 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
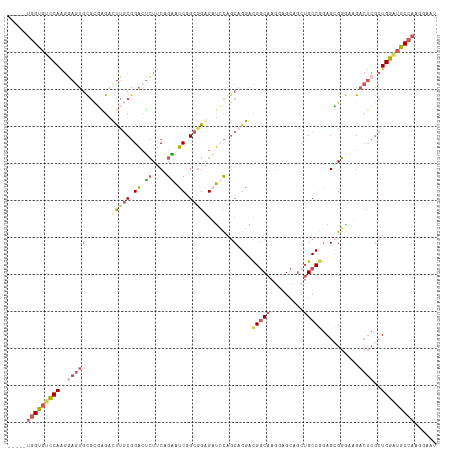
>dm3.chrX 3379943 111 + 22422827 -----UGGUGUUCAAGAAGUGCGCUAGAUUUGCGGACUCUUCAGAAUCGGCGGAGAUCCAGCAGGACGGCAAGGAGCAACUGCCUGAGAGGCAAGACUCGCUGGAUGCCAAGGAAU -----.(((....((((.((.(((.......))).))))))....)))(((....(((((((.....((((.(......)))))...(((......)))))))))))))....... ( -36.20, z-score = -0.33, R) >droSim1.chrX 2457411 111 + 17042790 -----UGGUGUCCAAGGAGUGCGCGAGACUUGCGGACUCUUCAGGAUCGGCGGGGAUCCAGCCGGACGGCAAGAAGCAGCUGCCGGAGCGGGAUGACUCGCUGGAUGCCAAGGAAU -----((((((((.((((((.(((((...))))).)))))).......((((((.((((..((((.((((........))))))))....))))..))))))))))))))...... ( -52.50, z-score = -3.09, R) >droSec1.super_4 2895177 111 - 6179234 -----UGGUGUCCAAGGAGUGCGCGAGACUUGCGGACUCUUCAGGAUCGGCGGGGAUCCAGCAGGACGGCAAGAAGCAGAUGCCGGAGCGGGAUGACUCGCUGGAUGCCAAGGAAU -----((((((((.((((((.(((((...))))).)))))).......((((((.((((.((....(((((.........)))))..)).))))..))))))))))))))...... ( -48.10, z-score = -2.89, R) >droYak2.chrX 16769657 114 - 21770863 --UGGUGGUGUCCAAGGAGUCUGGGGGAUCUGCGGAAUCUGCAGAAUCGGCAGCGCUGCAGCAGGACGGCAAGGAGCAGCUGCCGGAGCGGAAGGACUCGCUGGCAGCCAAGGAAU --...((((..(((..((((((......(((((((...))))))).(((((((((((.(.((......))..).))).)))))))).......))))))..)))..))))...... ( -46.70, z-score = -1.05, R) >droAna3.scaffold_13248 3633574 108 + 4840945 AGGGGAAGCGGCCAGCCAAGGCAUCUGCCU--CUGGCGGUCCUGCGUCCGGGGAGUCGUCACAGGAUGCCAAAGCGAAGCAGCCAAAGGGCGAAG------UGGCUGCAAAGGAAU .......(((((((((((((((....))))--.)))).(((((((((((...((....))...))))))....((......))...)))))....------)))))))........ ( -44.40, z-score = -0.69, R) >consensus _____UGGUGUCCAAGGAGUGCGCGAGACUUGCGGACUCUUCAGAAUCGGCGGAGAUCCAGCAGGACGGCAAGGAGCAGCUGCCGGAGCGGGAAGACUCGCUGGAUGCCAAGGAAU .....(((((((((..((((((......(((((.((.((....)).)).)))))......))....(((((.........)))))..........))))..)))))))))...... (-22.02 = -23.82 + 1.80)

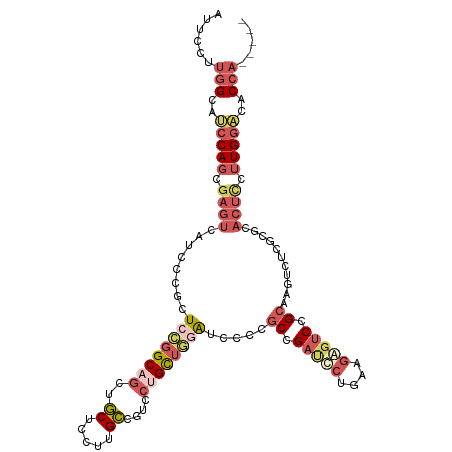
| Location | 3,379,943 – 3,380,054 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 70.68 |
| Shannon entropy | 0.52517 |
| G+C content | 0.61080 |
| Mean single sequence MFE | -38.10 |
| Consensus MFE | -15.72 |
| Energy contribution | -18.40 |
| Covariance contribution | 2.68 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.668652 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
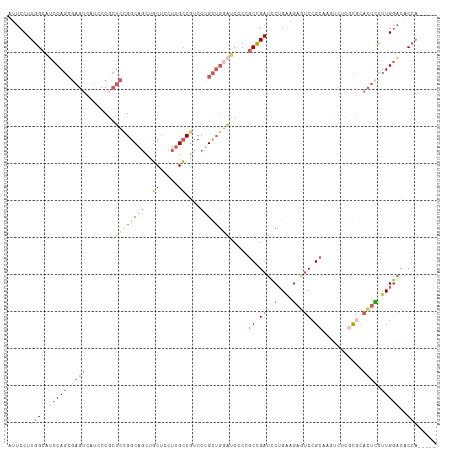
>dm3.chrX 3379943 111 - 22422827 AUUCCUUGGCAUCCAGCGAGUCUUGCCUCUCAGGCAGUUGCUCCUUGCCGUCCUGCUGGAUCUCCGCCGAUUCUGAAGAGUCCGCAAAUCUAGCGCACUUCUUGAACACCA----- .......((((...(((((...(((((.....))))))))))...)))).....(((((((....((.(((((....))))).))..))))))).................----- ( -33.30, z-score = -1.29, R) >droSim1.chrX 2457411 111 - 17042790 AUUCCUUGGCAUCCAGCGAGUCAUCCCGCUCCGGCAGCUGCUUCUUGCCGUCCGGCUGGAUCCCCGCCGAUCCUGAAGAGUCCGCAAGUCUCGCGCACUCCUUGGACACCA----- .....((((((((((((((((......))))((((((.......))))))....)))))))....)))))(((....((((.(((.......))).))))...))).....----- ( -37.80, z-score = -1.13, R) >droSec1.super_4 2895177 111 + 6179234 AUUCCUUGGCAUCCAGCGAGUCAUCCCGCUCCGGCAUCUGCUUCUUGCCGUCCUGCUGGAUCCCCGCCGAUCCUGAAGAGUCCGCAAGUCUCGCGCACUCCUUGGACACCA----- .....((((((((((((((((......))))(((((.........)))))....)))))))....)))))(((....((((.(((.......))).))))...))).....----- ( -37.60, z-score = -2.21, R) >droYak2.chrX 16769657 114 + 21770863 AUUCCUUGGCUGCCAGCGAGUCCUUCCGCUCCGGCAGCUGCUCCUUGCCGUCCUGCUGCAGCGCUGCCGAUUCUGCAGAUUCCGCAGAUCCCCCAGACUCCUUGGACACCACCA-- ......(((.((((((.(((((.........(((((((.(((.(..((......)).).))))))))))..(((((.......))))).......))))).)))).)))))...-- ( -40.70, z-score = -2.35, R) >droAna3.scaffold_13248 3633574 108 - 4840945 AUUCCUUUGCAGCCA------CUUCGCCCUUUGGCUGCUUCGCUUUGGCAUCCUGUGACGACUCCCCGGACGCAGGACCGCCAG--AGGCAGAUGCCUUGGCUGGCCGCUUCCCCU ........(((((((------..........)))))))...((((((((.(((((((.((......))..)))))))..)))))--)(((....((....))..)))))....... ( -41.10, z-score = -1.78, R) >consensus AUUCCUUGGCAUCCAGCGAGUCAUCCCGCUCCGGCAGCUGCUCCUUGCCGUCCUGCUGGAUCCCCGCCGAUCCUGAAGAGUCCGCAAGUCUCGCGCACUCCUUGGACACCA_____ .......((..(((((.((((........((((((((..((.....))....)))))))).....((.(((((....))))).))...........)))).)))))..))...... (-15.72 = -18.40 + 2.68)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:11:32 2011