
| Sequence ID | dm3.chrX |
|---|---|
| Location | 3,369,040 – 3,369,142 |
| Length | 102 |
| Max. P | 0.528978 |

| Location | 3,369,040 – 3,369,142 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 55.76 |
| Shannon entropy | 0.82906 |
| G+C content | 0.56442 |
| Mean single sequence MFE | -39.26 |
| Consensus MFE | -11.50 |
| Energy contribution | -12.95 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.82 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.29 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.528978 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
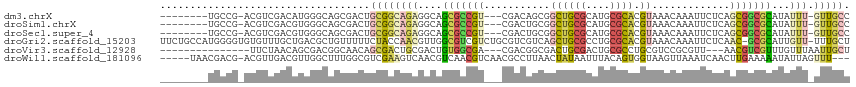
>dm3.chrX 3369040 102 - 22422827 --------UGCCG-ACGUCGACAUGGGCAGCGACUGCGGCAGAGGCAGCGCCGU---CGACAGCGGCUGCGCAUGCGCACGUAAACAAAUUCUCAGCGGCGCAUAUUU-GUUGCC --------(((((-..((((.(.......)))))..)))))..((((((((.((---((....)))).))))((((((.(((.............)))))))))....-..)))) ( -41.92, z-score = -0.42, R) >droSim1.chrX 2444844 102 - 17042790 --------UGCCG-ACGUCGACGUGGGCAGCGACUGCGGCAGAGGCAGCGCCGU---CGACUGCGGCUGCGCAUGCGCACGUAAACAAAUUCUCAGCGGCGCAUAUUU-GUUGCC --------((((.-(((....))).))))(((((.(((.(((..((((((....---)).))))..))))))((((((.(((.............)))))))))....-))))). ( -43.92, z-score = -0.37, R) >droSec1.super_4 2884415 102 + 6179234 --------UGCCG-ACGUCGACGUGGGCAGCGACUGCGGCAGAGGCAGCGCCGU---CGACUGCGGCUGCGCAUGCGCACGUAAACAAAUUCUCAGCGGCGCAUAUUU-GUUGCC --------((((.-(((....))).))))(((((.(((.(((..((((((....---)).))))..))))))((((((.(((.............)))))))))....-))))). ( -43.92, z-score = -0.37, R) >droGri2.scaffold_15203 11653731 113 + 11997470 UUCUGCCAUGGGGUGUGUUUGCUGACGCUGUUUUUCUACCAACGUUGGCGUCGUCUGCGUCGUCAGCUGCGCCUGCGCACGUAAACAAAUUCUCAAC-GCGCAUUGUU-UUUGCU ....((..(((((..(((((((((.(((.(((........)))(((((((.((....)).))))))).......))))).)))))))...)))))..-))(((.....-..))). ( -33.90, z-score = -0.11, R) >droVir3.scaffold_12928 2419432 94 - 7717345 ---------------UUCUAACAGCGACGGCAACAGCGACUGCGACUGUGGCGA---CGACGGCGACUGCGACUGCGCCUGCGUCCGCGUU---AACGUCGUUUGUUUAAUUGCU ---------------....((((((((((((.((((((....)).)))).))((---((.(((((...(((....)))...)).)))))))---..))))).)))))........ ( -33.30, z-score = -0.20, R) >droWil1.scaffold_181096 2034562 106 + 12416693 -----UAACGACG-ACGUUGACGUUGGCUUUGGCGUCGAAGUCAACGUCAACGUCAACGCCUUAACUAUAAUUUACAGUGGUAAGUUAAAUCAACUUGAAAAAUAUUAGUUU--- -----...((..(-((((((((((((((((((....)))))))))))))))))))..))....(((((((((((((....))))))))..((.....)).......))))).--- ( -38.60, z-score = -5.57, R) >consensus ________UGCCG_ACGUCGACGUGGGCAGCGACUGCGGCAGAGGCAGCGCCGU___CGACUGCGGCUGCGCAUGCGCACGUAAACAAAUUCUCAGCGGCGCAUAUUU_GUUGCC ........(((((...((((..........))))..)))))......(((((((...........((((((....)))).)).............)))))))............. (-11.50 = -12.95 + 1.45)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:11:31 2011