
| Sequence ID | dm3.chrX |
|---|---|
| Location | 3,343,454 – 3,343,546 |
| Length | 92 |
| Max. P | 0.997282 |

| Location | 3,343,454 – 3,343,546 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 65.55 |
| Shannon entropy | 0.56027 |
| G+C content | 0.44821 |
| Mean single sequence MFE | -34.80 |
| Consensus MFE | -21.27 |
| Energy contribution | -21.15 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.07 |
| SVM RNA-class probability | 0.997282 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 3343454 92 + 22422827 UUUGGCUCGACUGCUGGUGGGCCAAGGGCACAAUUGUCGAGUGUGCAAAUGAUGUACGUAUGUAUGUACA---AUGUACA--------------------ACGAACGACGACUAA (((((((((.(....).))))))))).......((((((.(((((((.....(((((((....)))))))---.))))).--------------------))...)))))).... ( -27.70, z-score = -1.27, R) >droAna3.scaffold_13334 6805 105 + 1562580 UUUAGUGUUCCAAGUUGAGGGCGAAGGGCA-ACUUGGAAUGUGUGCAAAAAAAAUGAAAAAAAAAACA-----AUAUUUUUGCGUCACUUGGAAAUUU---UUUACGAUGACAG- ....(..((((((((((....(....).))-))))))))..)..(((((((.(.((..........))-----.).)))))))((((.(((.......---....)))))))..- ( -24.40, z-score = -1.85, R) >droSec1.super_4 2859694 115 - 6179234 UUUGGCUCGACUGGUGGAGGGCCAAGGGCACGAUUGUCGAGUGUGCAAAUGAUGUACGUAUGUGUACAUACAUAUGUACAUGCGUCACUCGCUGUUUCGAACGAACGACGACUAA ((((((((..(....)..)))))))).......((((((((((((((.....((((((((((((....)))))))))))))))).)))))...((((.....))))))))).... ( -42.30, z-score = -2.64, R) >droSim1.chrX_random 1456272 115 + 5698898 UUUGGCUCGACUGGUGGAGGGCCAAGGGCACGCUUGUCGAGUGUGCAAAUGAUGUACGUAUGUGUACAUACAUAUGUACAUGCGUCACUCGCUGUUUCCAACGAACGACGACUAA ((((((((..(....)..)))))))).((((((((...)))))))).....(((((((((((((....))))))))))))).((((..(((.((....)).)))..))))..... ( -44.80, z-score = -3.11, R) >consensus UUUGGCUCGACUGGUGGAGGGCCAAGGGCACAAUUGUCGAGUGUGCAAAUGAUGUACGUAUGUAUACAUA___AUGUACAUGCGUCACUCGCUGUUUC__ACGAACGACGACUAA .((((((((((.((((....((.....)).)))).))))))).........((((((((((((......))))))))))))...........................))).... (-21.27 = -21.15 + -0.12)

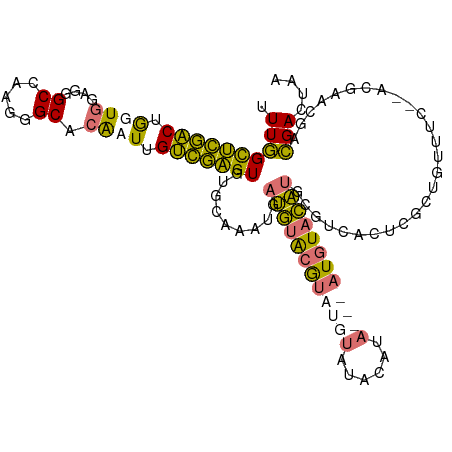
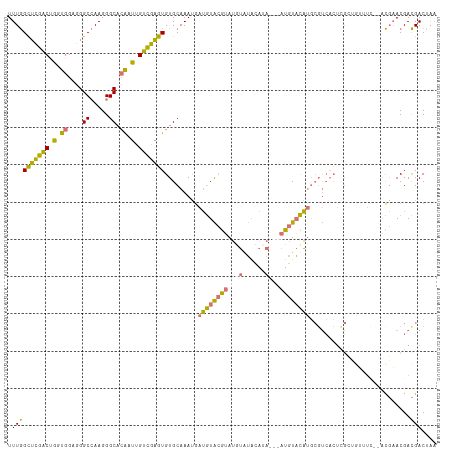
| Location | 3,343,454 – 3,343,546 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 65.55 |
| Shannon entropy | 0.56027 |
| G+C content | 0.44821 |
| Mean single sequence MFE | -27.64 |
| Consensus MFE | -10.40 |
| Energy contribution | -9.09 |
| Covariance contribution | -1.31 |
| Combinations/Pair | 1.60 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.916241 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 3343454 92 - 22422827 UUAGUCGUCGUUCGU--------------------UGUACAU---UGUACAUACAUACGUACAUCAUUUGCACACUCGACAAUUGUGCCCUUGGCCCACCAGCAGUCGAGCCAAA ......(((((..((--------------------(((((..---.))))).))..))).))............((((((..(((.(((...)))....)))..))))))..... ( -18.30, z-score = -0.27, R) >droAna3.scaffold_13334 6805 105 - 1562580 -CUGUCAUCGUAAA---AAAUUUCCAAGUGACGCAAAAAUAU-----UGUUUUUUUUUUCAUUUUUUUUGCACACAUUCCAAGU-UGCCCUUCGCCCUCAACUUGGAACACUAAA -.............---.........((((..(((((((...-----((..........))...))))))).....((((((((-((...........))))))))))))))... ( -21.20, z-score = -3.25, R) >droSec1.super_4 2859694 115 + 6179234 UUAGUCGUCGUUCGUUCGAAACAGCGAGUGACGCAUGUACAUAUGUAUGUACACAUACGUACAUCAUUUGCACACUCGACAAUCGUGCCCUUGGCCCUCCACCAGUCGAGCCAAA ...(.(((((((((((......)))))))))))).(((.((.(((.((((((......))))))))).)).)))((((((......(((...))).........))))))..... ( -31.66, z-score = -1.82, R) >droSim1.chrX_random 1456272 115 - 5698898 UUAGUCGUCGUUCGUUGGAAACAGCGAGUGACGCAUGUACAUAUGUAUGUACACAUACGUACAUCAUUUGCACACUCGACAAGCGUGCCCUUGGCCCUCCACCAGUCGAGCCAAA ...(.((((((((((((....)))))))))))))((((((.(((((......))))).)))))).....((((.((.....)).))))..(((((.(..(....)..).))))). ( -39.40, z-score = -3.37, R) >consensus UUAGUCGUCGUUCGU__GAAACAGCGAGUGACGCAUGUACAU___UAUGUACACAUACGUACAUCAUUUGCACACUCGACAAGCGUGCCCUUGGCCCUCCACCAGUCGAGCCAAA ...................................(((((((....))))))).....................((((((......((.....)).........))))))..... (-10.40 = -9.09 + -1.31)

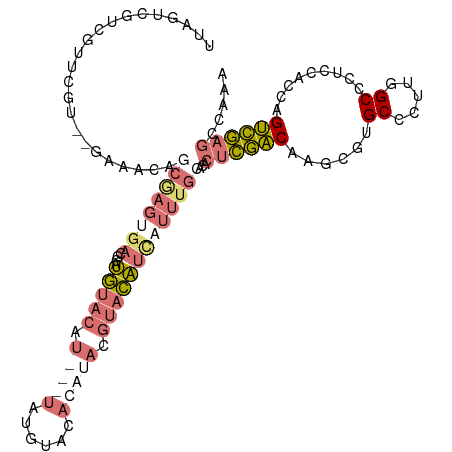
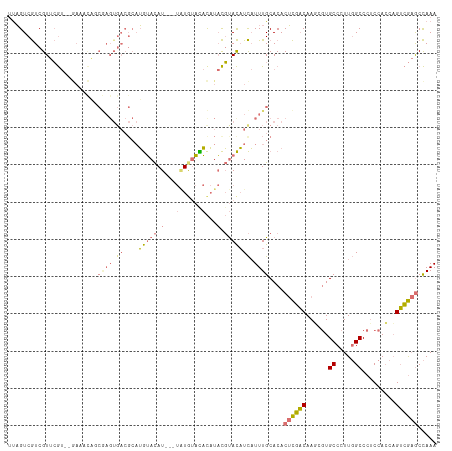
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:11:29 2011