
| Sequence ID | dm3.chrX |
|---|---|
| Location | 3,321,474 – 3,321,566 |
| Length | 92 |
| Max. P | 0.993539 |

| Location | 3,321,474 – 3,321,566 |
|---|---|
| Length | 92 |
| Sequences | 11 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.75 |
| Shannon entropy | 0.46451 |
| G+C content | 0.45996 |
| Mean single sequence MFE | -26.35 |
| Consensus MFE | -25.01 |
| Energy contribution | -24.38 |
| Covariance contribution | -0.63 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.95 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.62 |
| SVM RNA-class probability | 0.993539 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
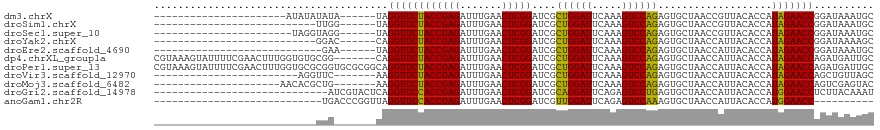
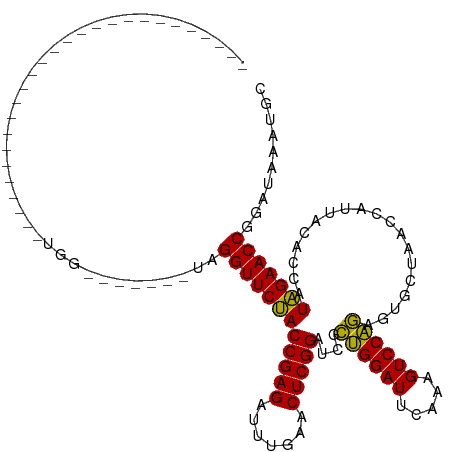
>dm3.chrX 3321474 92 + 22422827 ----------------------AUAUAUAUA------UAGGUUCUACCGAGAUUUGAACUCGGAUCGCUGGAUUCAAAGUCCAGAGUGCUAACCGUUACACCAUAGAACCGGAUAAAUGC ----------------------.(((.(((.------..((((((((((((.......)))))....((((((.....))))))...................)))))))..))).))). ( -24.80, z-score = -2.53, R) >droSim1.chrX 2401547 87 + 17042790 ---------------------------UUGG------UAGGUUCUACCGAGAUUUGAACUCGGAUCGCUGGAUUCAAAGUCCAGAGUGCUAACCGUUACACCAUAGAACCGGAUAAAUGC ---------------------------....------..((((((((((((.......)))))....((((((.....))))))...................))))))).......... ( -24.40, z-score = -1.78, R) >droSec1.super_10 3020653 91 + 3036183 -----------------------UAGGUAGG------UAGGUUCUACCGAGAUUUGAACUCGGAUCGCUGGAUUCAAAGUCCAGAGUGCUAACCGUUACACCAUAGAACCGGAUAAAUGC -----------------------........------..((((((((((((.......)))))....((((((.....))))))...................))))))).......... ( -24.40, z-score = -1.26, R) >droYak2.chrX 16710371 87 - 21770863 ---------------------------GGAC------CAGGUUCUACCGAGAUUUGAACUCGGAUCGCUGGAUUCAAAGUCCAGAGUGCUAACCAUUACACCAUAGAACCGGAUAAAAGC ---------------------------...(------(.((((((((((((.......)))))....((((((.....))))))...................)))))))))........ ( -26.00, z-score = -2.91, R) >droEre2.scaffold_4690 729216 86 + 18748788 ----------------------------GAA------UAGGUUCUACCGAGAUUUGAACUCGGAUCGCUGGAUUCAAAGUCCAGAGUGCUAACCAUUACACCAUAGAACCGGAUAAAUGC ----------------------------...------..((((((((((((.......)))))....((((((.....))))))...................))))))).......... ( -24.40, z-score = -2.91, R) >dp4.chrXL_group1a 7445405 113 + 9151740 CGUAAAGUAUUUUCGAACUUUGGUGUGCGG-------CAGGUUCUACCGAGAUUUGAACUCGGAUCGCUGGAUUCAAAGUCCAGAGUGCUAACCAUUACACCAUAGAACCAGAUGAUUGC .((((..(((((.....((.(((((((.((-------..(((.((.(((((.......)))))....((((((.....)))))))).)))..))..))))))).))....))))).)))) ( -30.10, z-score = -1.14, R) >droPer1.super_13 2086159 120 + 2293547 CGUAAAGUAUUUUCGAACUUUGGUGCGCGGUGCGCGGCAGGUUCUACCGAGAUUUGAACUCGGAUCGCUGGAUUCAAAGUCCAGAGUGCUAACCAUUACACCAUAGAACCAGAUGAUUGC .((((..(((((........((.(((((...))))).))((((((((((((.......)))))....((((((.....))))))...................)))))))))))).)))) ( -33.80, z-score = -0.83, R) >droVir3.scaffold_12970 3786034 89 - 11907090 ------------------------AGGUUC-------AAGGUUCUACCGAGAUUUGAACUCGGAUCGCUGGAUUCAAAGUCCAGAGUGCUAACCAUUACACCAUAGAACCAGCUGUUAGC ------------------------.((((.-------..((((((((((((.......)))))....((((((.....))))))...................)))))))))))...... ( -24.50, z-score = -1.83, R) >droMoj3.scaffold_6482 1310530 92 + 2735782 ---------------------AACACGCUG-------AAGGUUCUACCGAGAUUUGAACUCGGAUCGCUGGAUUCAAAGUCCAGAGUGCUAACCAUUACACCAUAGAACCAGUCGAGUAC ---------------------.....((((-------(.((((((((((((.......)))))....((((((.....))))))...................)))))))..)).))).. ( -25.90, z-score = -2.37, R) >droGri2.scaffold_14978 67051 91 - 1124632 -----------------------------AUCGUACUCAGGUUCCACCGAGAUUUGAACUCGGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUGGAACCUCUUACAAAU -----------------------------...(((...(((((((((((((.......))))).....((.((((((....)))))).)).............))))))))..))).... ( -28.90, z-score = -3.41, R) >anoGam1.chr2R 59258898 82 + 62725911 ----------------------------UGACCCGGUUAGGUUCCACCGAGAUUUGAACUCGGAUCGUUGGAUUCAGAGUCCAAAGUGCUAACCAUUACACCAUGGAACC---------- ----------------------------...........((((((((((((.......)))))....((((((.....))))))...................)))))))---------- ( -22.70, z-score = -1.22, R) >consensus ___________________________UGG_______UAGGUUCUACCGAGAUUUGAACUCGGAUCGCUGGAUUCAAAGUCCAGAGUGCUAACCAUUACACCAUAGAACCGGAUAAAUGC .......................................((((((((((((.......)))))....((((((.....))))))...................))))))).......... (-25.01 = -24.38 + -0.63)

| Location | 3,321,474 – 3,321,566 |
|---|---|
| Length | 92 |
| Sequences | 11 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.75 |
| Shannon entropy | 0.46451 |
| G+C content | 0.45996 |
| Mean single sequence MFE | -25.69 |
| Consensus MFE | -24.41 |
| Energy contribution | -23.78 |
| Covariance contribution | -0.63 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.95 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.977046 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
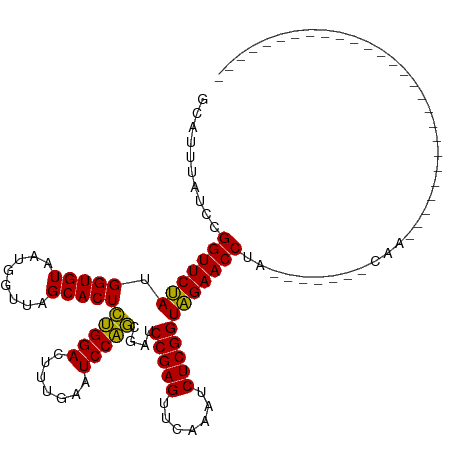
>dm3.chrX 3321474 92 - 22422827 GCAUUUAUCCGGUUCUAUGGUGUAACGGUUAGCACUCUGGACUUUGAAUCCAGCGAUCCGAGUUCAAAUCUCGGUAGAACCUA------UAUAUAUAU---------------------- ..........(((((((.(((((........)))))(((((.......)))))....(((((.......))))))))))))..------.........---------------------- ( -23.70, z-score = -1.65, R) >droSim1.chrX 2401547 87 - 17042790 GCAUUUAUCCGGUUCUAUGGUGUAACGGUUAGCACUCUGGACUUUGAAUCCAGCGAUCCGAGUUCAAAUCUCGGUAGAACCUA------CCAA--------------------------- ..........(((((((.(((((........)))))(((((.......)))))....(((((.......))))))))))))..------....--------------------------- ( -23.70, z-score = -1.62, R) >droSec1.super_10 3020653 91 - 3036183 GCAUUUAUCCGGUUCUAUGGUGUAACGGUUAGCACUCUGGACUUUGAAUCCAGCGAUCCGAGUUCAAAUCUCGGUAGAACCUA------CCUACCUA----------------------- ..........(((((((.(((((........)))))(((((.......)))))....(((((.......))))))))))))..------........----------------------- ( -23.70, z-score = -1.40, R) >droYak2.chrX 16710371 87 + 21770863 GCUUUUAUCCGGUUCUAUGGUGUAAUGGUUAGCACUCUGGACUUUGAAUCCAGCGAUCCGAGUUCAAAUCUCGGUAGAACCUG------GUCC--------------------------- ........(((((((((.(((((........)))))(((((.......)))))....(((((.......)))))))))))).)------)...--------------------------- ( -24.80, z-score = -1.53, R) >droEre2.scaffold_4690 729216 86 - 18748788 GCAUUUAUCCGGUUCUAUGGUGUAAUGGUUAGCACUCUGGACUUUGAAUCCAGCGAUCCGAGUUCAAAUCUCGGUAGAACCUA------UUC---------------------------- ..........(((((((.(((((........)))))(((((.......)))))....(((((.......))))))))))))..------...---------------------------- ( -23.70, z-score = -1.90, R) >dp4.chrXL_group1a 7445405 113 - 9151740 GCAAUCAUCUGGUUCUAUGGUGUAAUGGUUAGCACUCUGGACUUUGAAUCCAGCGAUCCGAGUUCAAAUCUCGGUAGAACCUG-------CCGCACACCAAAGUUCGAAAAUACUUUACG ...........(..((.((((((...((((.(.((((.((((.(((....))).).))))))).).)))).((((((...)))-------))).)))))).))..).............. ( -26.20, z-score = -0.64, R) >droPer1.super_13 2086159 120 - 2293547 GCAAUCAUCUGGUUCUAUGGUGUAAUGGUUAGCACUCUGGACUUUGAAUCCAGCGAUCCGAGUUCAAAUCUCGGUAGAACCUGCCGCGCACCGCGCACCAAAGUUCGAAAAUACUUUACG (((.......(((((((.(((((........)))))(((((.......)))))....(((((.......))))))))))))))).((((...))))...(((((........)))))... ( -30.71, z-score = -0.81, R) >droVir3.scaffold_12970 3786034 89 + 11907090 GCUAACAGCUGGUUCUAUGGUGUAAUGGUUAGCACUCUGGACUUUGAAUCCAGCGAUCCGAGUUCAAAUCUCGGUAGAACCUU-------GAACCU------------------------ ((.....)).(((((((.(((((........)))))(((((.......)))))....(((((.......))))))))))))..-------......------------------------ ( -24.10, z-score = -1.24, R) >droMoj3.scaffold_6482 1310530 92 - 2735782 GUACUCGACUGGUUCUAUGGUGUAAUGGUUAGCACUCUGGACUUUGAAUCCAGCGAUCCGAGUUCAAAUCUCGGUAGAACCUU-------CAGCGUGUU--------------------- ..((.((...(((((((.(((((........)))))(((((.......)))))....(((((.......))))))))))))..-------...)).)).--------------------- ( -25.40, z-score = -1.00, R) >droGri2.scaffold_14978 67051 91 + 1124632 AUUUGUAAGAGGUUCCAUGGUGUAAUGGUUAGCACUCAGGACUCUGAAUCCUGCGAUCCGAGUUCAAAUCUCGGUGGAACCUGAGUACGAU----------------------------- ..(((((..((((((((.(((((........)))))(((((.......)))))....(((((.......)))))))))))))...))))).----------------------------- ( -30.20, z-score = -2.31, R) >anoGam1.chr2R 59258898 82 - 62725911 ----------GGUUCCAUGGUGUAAUGGUUAGCACUUUGGACUCUGAAUCCAACGAUCCGAGUUCAAAUCUCGGUGGAACCUAACCGGGUCA---------------------------- ----------(((((((((((((........)))))((((((((.((.((....)))).))))))))......))))))))...........---------------------------- ( -26.40, z-score = -1.55, R) >consensus GCAUUUAUCCGGUUCUAUGGUGUAAUGGUUAGCACUCUGGACUUUGAAUCCAGCGAUCCGAGUUCAAAUCUCGGUAGAACCUA_______CAA___________________________ ..........(((((((.(((((........)))))(((((.......)))))....(((((.......))))))))))))....................................... (-24.41 = -23.78 + -0.63)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:11:26 2011