
| Sequence ID | dm3.chrX |
|---|---|
| Location | 3,274,420 – 3,274,538 |
| Length | 118 |
| Max. P | 0.788681 |

| Location | 3,274,420 – 3,274,538 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 73.14 |
| Shannon entropy | 0.43584 |
| G+C content | 0.53935 |
| Mean single sequence MFE | -45.92 |
| Consensus MFE | -25.09 |
| Energy contribution | -28.40 |
| Covariance contribution | 3.31 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.788681 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
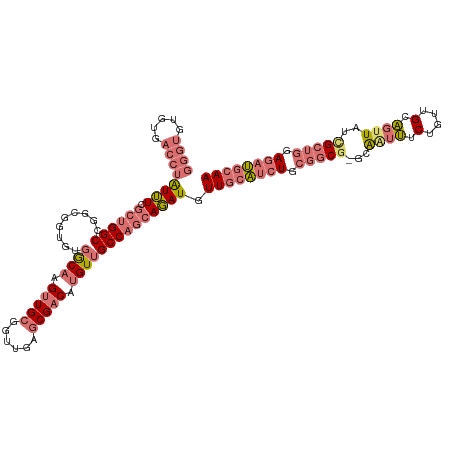
>dm3.chrX 3274420 118 + 22422827 GGGUGUGUGACCUAUUUCGCUGGCCGUUGGUGUGGCAAGUUGCGGUUGAGCGACAUGUUGCCAGCAGAUGUUGCAUCUGCGGCU-GCAAUUUCUGUUGCAGUUAUCGCUGUAGAUGCAA .(((..((((......))))..)))((((((..((((.(((((......))))).)))))))))).....((((((((((((((-(((((....))))))......))))))))))))) ( -49.80, z-score = -2.56, R) >droSim1.chrX 2360484 118 + 17042790 GGGUGUGUGACCUAUUUCGCUGGCCGGCGGUGUGGCAAGUUGCGGUUGAGCGACAUGUUGCCAGCAGAUGUUGCAUCUGCGGCG-GCAAUUUCUGUUGCAGCUAUCGCUGGAGAUGCAA ((((.....)))).....((...((((((((((.(((((((((......))))).(((((((.(((((((...)))))))))))-))).......)))).)).))))))))....)).. ( -51.80, z-score = -2.05, R) >droSec1.super_10 2979438 118 + 3036183 GGGUGUGUGACCUAUUUCGCUGGCCGGCGGUGUGGCAAGUUGCGGUUGAGCGACAUGUUGCCAGCAGAUGUUGCAUCUGCGGCG-GCAAUUUCUGUUGCAGUUAUCGCUGGAGAUGCAA ((((.....)))).....((...(((((((((..(((((((((......))))).(((((((.(((((((...)))))))))))-))).......))))...)))))))))....)).. ( -51.90, z-score = -2.40, R) >dp4.chrXL_group1a 7390155 103 + 9151740 ------------AGUGUCUGGGGCUGCUGGCU--ACCGGCUGAGA--GAGCGACAUGUUGCCGACACAUGUUGCUAGUGCAACGAGGGGCGUCUUUUGACCUUUUUGCAAAAAAAAAAA ------------...(.(((((((.....)))--.))))).....--.((((((((((.......))))))))))..(((((.((((..((.....)).)))).))))).......... ( -30.20, z-score = 0.12, R) >consensus GGGUGUGUGACCUAUUUCGCUGGCCGGCGGUGUGGCAAGUUGCGGUUGAGCGACAUGUUGCCAGCAGAUGUUGCAUCUGCGGCG_GCAAUUUCUGUUGCAGUUAUCGCUGGAGAUGCAA ((((.....))))((((.((((((.........((((.(((((......))))).)))))))))))))).((((((((.(((((.(((((....)))))......))))).)))))))) (-25.09 = -28.40 + 3.31)


| Location | 3,274,420 – 3,274,538 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 73.14 |
| Shannon entropy | 0.43584 |
| G+C content | 0.53935 |
| Mean single sequence MFE | -31.77 |
| Consensus MFE | -20.02 |
| Energy contribution | -21.03 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.742578 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 3274420 118 - 22422827 UUGCAUCUACAGCGAUAACUGCAACAGAAAUUGC-AGCCGCAGAUGCAACAUCUGCUGGCAACAUGUCGCUCAACCGCAACUUGCCACACCAACGGCCAGCGAAAUAGGUCACACACCC .((.(((((.(((((((..(((((......))))-)((((((((((...))))))).)))....)))))))....(((.....(((........)))..)))...)))))))....... ( -35.90, z-score = -2.79, R) >droSim1.chrX 2360484 118 - 17042790 UUGCAUCUCCAGCGAUAGCUGCAACAGAAAUUGC-CGCCGCAGAUGCAACAUCUGCUGGCAACAUGUCGCUCAACCGCAACUUGCCACACCGCCGGCCAGCGAAAUAGGUCACACACCC ((((.(((.((((....))))....)))....((-((.((((((((...)))))))((((((...((.((......)).))))))))....).))))..))))....(((.....))). ( -34.60, z-score = -1.24, R) >droSec1.super_10 2979438 118 - 3036183 UUGCAUCUCCAGCGAUAACUGCAACAGAAAUUGC-CGCCGCAGAUGCAACAUCUGCUGGCAACAUGUCGCUCAACCGCAACUUGCCACACCGCCGGCCAGCGAAAUAGGUCACACACCC ((((......(((((((...((((......))))-.((((((((((...))))))).)))....))))))).....))))((.(((........))).)).......(((.....))). ( -33.50, z-score = -1.45, R) >dp4.chrXL_group1a 7390155 103 - 9151740 UUUUUUUUUUUGCAAAAAGGUCAAAAGACGCCCCUCGUUGCACUAGCAACAUGUGUCGGCAACAUGUCGCUC--UCUCAGCCGGU--AGCCAGCAGCCCCAGACACU------------ .........((((......(((....)))((..((.((((....(((.((((((.......)))))).))).--...)))).)).--.))..))))...........------------ ( -23.10, z-score = 0.04, R) >consensus UUGCAUCUCCAGCGAUAACUGCAACAGAAAUUGC_CGCCGCAGAUGCAACAUCUGCUGGCAACAUGUCGCUCAACCGCAACUUGCCACACCACCGGCCAGCGAAAUAGGUCACACACCC ((((.......((((((...((((......))))..((((((((((...))))))).)))....)))))).............(((........)))..))))................ (-20.02 = -21.03 + 1.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:11:21 2011