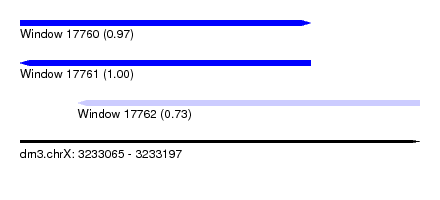
| Sequence ID | dm3.chrX |
|---|---|
| Location | 3,233,065 – 3,233,197 |
| Length | 132 |
| Max. P | 0.997367 |

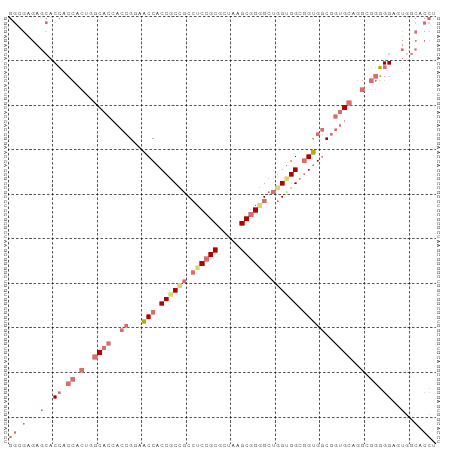
| Location | 3,233,065 – 3,233,161 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 66.84 |
| Shannon entropy | 0.55344 |
| G+C content | 0.73798 |
| Mean single sequence MFE | -47.73 |
| Consensus MFE | -29.04 |
| Energy contribution | -33.10 |
| Covariance contribution | 4.06 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.969753 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
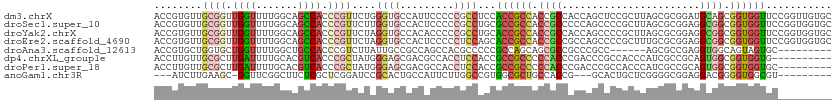
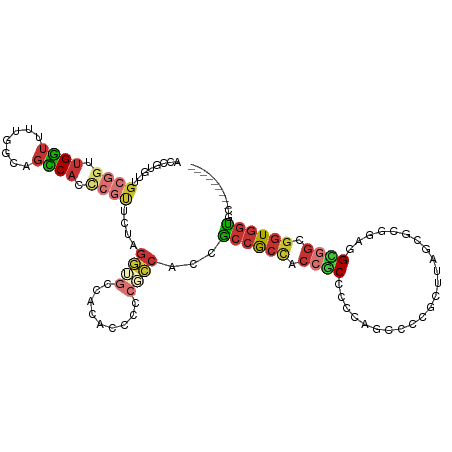
>dm3.chrX 3233065 96 + 22422827 GGCGAGAGCACCACCACAGGCACAACCGGAACCACCGCUGCAUCCGCGCUAAGCGGAGCUGGUGGCGGUGGCGGUGGAGGCGGGGGAAUGGCACCC .((....)).(((((...((.....))....(((((((..(.(((((.....)))))....)..))))))).))))).....(((........))) ( -46.00, z-score = -1.74, R) >droYak2.chrX 16623226 96 - 21770863 GGGGAGAGCACCACCACUGGCACCACCGGAACCACCGCCGCCUCCGCGCUAAGCGGGGCUGGUGGCGGUGGCGGUGCAGGCGGGGGUGUGGCACCU (((....(((((.((.((.(((((.((...(((.((((((((.((((.....))))))).))))).))))).))))).)).)).)))))....))) ( -56.40, z-score = -1.95, R) >droEre2.scaffold_4690 644341 96 + 18748788 GGCGAGAGCACCACCACUGGCACCACCGGAACCACCGCCGCCUCCGCGCAAAGCGGGGCUGGCGGCGGUGGCGGUGCUGGAGGGGGAGUGGCACCU ((.(...((.((.((.(..(((((.((...(((.((((((((.((((.....))))))).))))).))))).)))))..).)).)).))..).)). ( -56.00, z-score = -2.49, R) >anoGam1.chr3R 22658757 78 + 53272125 ---------ACGGAGGGCGGCACCAAGGACGCCACCCCGUCCUCCGCCCCGAGCAGUGCCGGUGGCAGCGCCACGGCCAAGAAUGGC--------- ---------.(((.((((((.....((((((......)))))))))))).........)))(((((...))))).((((....))))--------- ( -32.50, z-score = 0.20, R) >consensus GGCGAGAGCACCACCACUGGCACCACCGGAACCACCGCCGCCUCCGCGCUAAGCGGGGCUGGUGGCGGUGGCGGUGCAGGCGGGGGAGUGGCACCU ..........((.((.((((((((.((...(((.((((((.((((((.....)))))).)))))).))))).)))))))).))))........... (-29.04 = -33.10 + 4.06)


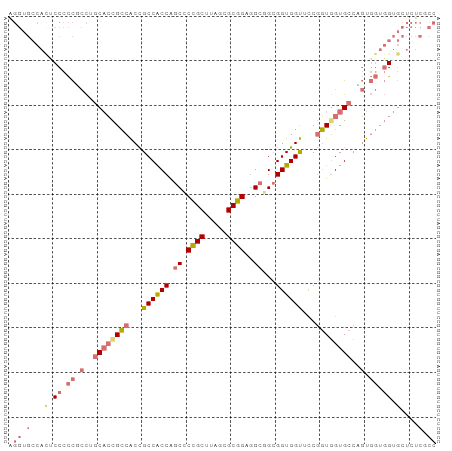
| Location | 3,233,065 – 3,233,161 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 66.84 |
| Shannon entropy | 0.55344 |
| G+C content | 0.73798 |
| Mean single sequence MFE | -49.20 |
| Consensus MFE | -30.71 |
| Energy contribution | -34.02 |
| Covariance contribution | 3.31 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.09 |
| SVM RNA-class probability | 0.997367 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 3233065 96 - 22422827 GGGUGCCAUUCCCCCGCCUCCACCGCCACCGCCACCAGCUCCGCUUAGCGCGGAUGCAGCGGUGGUUCCGGUUGUGCCUGUGGUGGUGCUCUCGCC (((..((((..(..(((..((...((((((((.....(((((((.....))))).)).))))))))...))..)))...)..))))..)))..... ( -43.60, z-score = -1.62, R) >droYak2.chrX 16623226 96 + 21770863 AGGUGCCACACCCCCGCCUGCACCGCCACCGCCACCAGCCCCGCUUAGCGCGGAGGCGGCGGUGGUUCCGGUGGUGCCAGUGGUGGUGCUCUCCCC .((.(...((((.((((..((((((((((((((.((.(((((((.....)))).))))).))))))...))))))))..)))).))))..).)).. ( -56.00, z-score = -3.26, R) >droEre2.scaffold_4690 644341 96 - 18748788 AGGUGCCACUCCCCCUCCAGCACCGCCACCGCCGCCAGCCCCGCUUUGCGCGGAGGCGGCGGUGGUUCCGGUGGUGCCAGUGGUGGUGCUCUCGCC .((((.(((..((.((...(((((((((((((((((.(((((((.....)))).))))))))))))...)))))))).)).))..)))....)))) ( -57.50, z-score = -3.75, R) >anoGam1.chr3R 22658757 78 - 53272125 ---------GCCAUUCUUGGCCGUGGCGCUGCCACCGGCACUGCUCGGGGCGGAGGACGGGGUGGCGUCCUUGGUGCCGCCCUCCGU--------- ---------((((....)))).(((((((((((.(((((...)).)))))).(((((((......))))))))))))))).......--------- ( -39.70, z-score = -0.33, R) >consensus AGGUGCCACUCCCCCGCCUGCACCGCCACCGCCACCAGCCCCGCUUAGCGCGGAGGCGGCGGUGGUUCCGGUGGUGCCAGUGGUGGUGCUCUCGCC .((((...((((.((((..((((((((...((((((.(((((((.....)))).)))...))))))...))))))))..)))).))))....)))) (-30.71 = -34.02 + 3.31)


| Location | 3,233,084 – 3,233,197 |
|---|---|
| Length | 113 |
| Sequences | 8 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 64.15 |
| Shannon entropy | 0.72083 |
| G+C content | 0.69881 |
| Mean single sequence MFE | -46.92 |
| Consensus MFE | -17.23 |
| Energy contribution | -18.31 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.55 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.734326 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 3233084 113 - 22422827 ACCGUGUUGCGGUUGGUUUUGGCAGCCACCCGUUCUGGGUGCCAUUCCCCCGCCUCCACCGCCACCGCCACCAGCUCCGCUUAGCGCGGAUGCAGCGGUGGUUCCGGUUGUGC ((((((..((((..((...(((((.(((.......))).)))))..)).))))...))).((((((((.....(((((((.....))))).)).))))))))...)))..... ( -49.90, z-score = -1.84, R) >droSec1.super_10 2938671 113 - 3036183 ACCGUGUUGCGGUUGGUUUUGGCAGCCACCCGUUCUUGGUGCCACUCCCCCGCCUGCGCCGCCACCGCCCCCAGCCCCGCUUAGCGCGGAGGCGGCGGUGGUUCCGGUGGUGC ........((((..((...(((((.(((........))))))))..)).))))..((((((((((((((.((.(((((((.....)))).))))).))))))...)))))))) ( -57.30, z-score = -1.86, R) >droYak2.chrX 16623245 113 + 21770863 ACCGUGUUGCGGUUGGUUUUGGCAGCCACCCGUUCUAGGUGCCACACCCCCGCCUGCACCGCCACCGCCACCAGCCCCGCUUAGCGCGGAGGCGGCGGUGGUUCCGGUGGUGC ........((((..(((..(((((.((..........))))))).))).))))..((((((((((((((.((.(((((((.....)))).))))).))))))...)))))))) ( -57.70, z-score = -2.05, R) >droEre2.scaffold_4690 644360 113 - 18748788 ACCGUGUUGCGGUUGGUUUUGGCAGCCACCCGUUCUAGGUGCCACUCCCCCUCCAGCACCGCCACCGCCGCCAGCCCCGCUUUGCGCGGAGGCGGCGGUGGUUCCGGUGGUGC (((.....((((.(((((.....))))).))))....)))...............(((((((((((((((((.(((((((.....)))).))))))))))))...)))))))) ( -60.20, z-score = -3.13, R) >droAna3.scaffold_12613 367086 98 + 519072 ACCGUGCUGGUGCUGGUUUUGGCUGCCACCCGUCUUAUUGCCGCCAGCCACGCCCCCGCCAGCAGCGCCGCCCGCC------AGCGCCGAGGUGGCAGUAGUGC--------- ....((((((((..(((..((((((.(...((......))..).)))))).)))..))))))))((((..(..(((------(.(.....).)))).)..))))--------- ( -39.80, z-score = 0.19, R) >dp4.chrXL_group1e 2776487 103 - 12523060 ACCUUGUUGCGCUUGAUUUUGCACGUCACCCGCUAUGGGAGCGACGCCACCUCCACCGCCGCCCCCACCCGACCCGCCACCCAUCGCCGCAGUGGCGGUGGUG---------- .....(((((((........))......(((.....))).)))))((((((.((((.((.((.......................)).)).)))).)))))).---------- ( -33.80, z-score = -0.63, R) >droPer1.super_18 1673607 104 - 1952607 ACCUUGUUGCGCUUGAUUUUGCACGUCACCCGCUAUGGGAGCGACGCCACCUCCACCGCCGCCCCCACCCGACCCGCCACCCAUCGCCGCAGUGGCGGUGGUGC--------- .....(((((((........))......(((.....))).)))))((((((.((((.((.((.......................)).)).)))).))))))..--------- ( -33.80, z-score = -0.15, R) >anoGam1.chr3R 22658776 97 - 53272125 ---AUCUUGAAGC-GGUUCGGCUUCUCGCUCGGAUCCGCACUGCCAUUCUUGGCCGUGGCGCUGCCACCG---GCACUGCUCGGGGCGGAGGACGGGGUGGCGU--------- ---........((-((((((((.....)).)))).))))((.((((....)))).)).((((..((.(((---.(.((((.....)))).)..)))))..))))--------- ( -42.90, z-score = -0.26, R) >consensus ACCGUGUUGCGGUUGGUUUUGGCAGCCACCCGUUCUAGGUGCCACACCCCCGCCACCGCCGCCACCGCCCCCAGCCCCGCUUAGCGCGGAGGCGGCGGUGGUGC_________ ........((((.((((.......)))).))))....((((.........))))...((((((.((((.......................)))).))))))........... (-17.23 = -18.31 + 1.08)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:11:17 2011