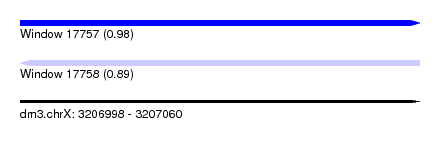
| Sequence ID | dm3.chrX |
|---|---|
| Location | 3,206,998 – 3,207,060 |
| Length | 62 |
| Max. P | 0.982184 |

| Location | 3,206,998 – 3,207,060 |
|---|---|
| Length | 62 |
| Sequences | 7 |
| Columns | 67 |
| Reading direction | forward |
| Mean pairwise identity | 64.79 |
| Shannon entropy | 0.69477 |
| G+C content | 0.49829 |
| Mean single sequence MFE | -10.07 |
| Consensus MFE | -4.99 |
| Energy contribution | -4.10 |
| Covariance contribution | -0.89 |
| Combinations/Pair | 2.14 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.10 |
| SVM RNA-class probability | 0.982184 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 3206998 62 + 22422827 CACUCGCAGCCAGAUAGACAAGCGUGU----CCAUCCAGCAGUUUAUCCGCCAGUUUGUCAGUCAC- ............(((.(((((((.((.----.....))((.(.....).))..))))))).)))..- ( -10.70, z-score = -1.37, R) >droEre2.scaffold_4690 617842 66 + 18748788 CACUCGCAGCCAGAUAGACAGAUAGACCAGUCCAUCCAGCAGUUUAUCCACCAGUUUGUCAGUCAC- ............(((((((.(((((((..((.......)).))))))).....)))))))......- ( -10.40, z-score = -2.29, R) >droYak2.chrX 16594763 64 - 21770863 CAUUCGCAGCCAGAUAGACAGAUAGAC--GUCCAUCCAGCAGUUUAUCCACCAGUUUGUCAGUCAC- ............(((((((.(((((((--............))))))).....)))))))......- ( -11.20, z-score = -3.17, R) >droSec1.super_10 2913797 60 + 3036183 CACUCGCAGCCACAUAGACAAGCGU------CCAUCCAGCAGUUUAUCCGCCAGUUUGUCAGUCAC- ................((((((((.------.....).((.(.....).))..)))))))......- ( -7.40, z-score = -1.05, R) >droSim1.chrX 2294123 60 + 17042790 CACUCGCAGCCAGAUAGACAAGCGU------CCAUCCAGCAGUUUAUCCGCCAGUUUGUCAGUCAC- ............(((.((((((((.------.....).((.(.....).))..))))))).)))..- ( -10.10, z-score = -1.99, R) >droGri2.scaffold_14853 4390204 63 + 10151454 CACUCAAAAGCCACUCAAUCGUCAAUU----CAGUCAAUCUGUGAAUCAAUCAGACUGCCAAUCAAU ...........................----(((((....((.....))....)))))......... ( -4.30, z-score = 0.17, R) >anoGam1.chr2R 42725602 63 - 62725911 AAAUCAACCUCGGACGGAUAUGCGUCGU--GCGGUUAGGC-GUGUGUGUGUGUGUGUAACAAUGAG- ........((((.(((.(((((((.((.--.((......)-)..))))))))).))).....))))- ( -16.40, z-score = -0.26, R) >consensus CACUCGCAGCCAGAUAGACAAGCGU______CCAUCCAGCAGUUUAUCCGCCAGUUUGUCAGUCAC_ ................(((((((..............................)))))))....... ( -4.99 = -4.10 + -0.89)

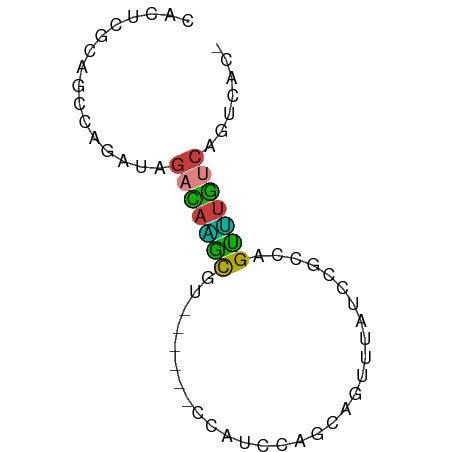
| Location | 3,206,998 – 3,207,060 |
|---|---|
| Length | 62 |
| Sequences | 7 |
| Columns | 67 |
| Reading direction | reverse |
| Mean pairwise identity | 64.79 |
| Shannon entropy | 0.69477 |
| G+C content | 0.49829 |
| Mean single sequence MFE | -16.00 |
| Consensus MFE | -4.44 |
| Energy contribution | -4.39 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.77 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.28 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.891910 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
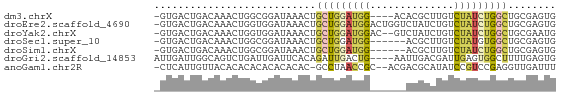
>dm3.chrX 3206998 62 - 22422827 -GUGACUGACAAACUGGCGGAUAAACUGCUGGAUGG----ACACGCUUGUCUAUCUGGCUGCGAGUG -...........(((.(((......).((..(((((----(((....))))))))..)).)).))). ( -21.50, z-score = -3.97, R) >droEre2.scaffold_4690 617842 66 - 18748788 -GUGACUGACAAACUGGUGGAUAAACUGCUGGAUGGACUGGUCUAUCUGUCUAUCUGGCUGCGAGUG -...........(((.(((......).((..(((((((.((....)).)))))))..)).)).))). ( -18.10, z-score = -2.38, R) >droYak2.chrX 16594763 64 + 21770863 -GUGACUGACAAACUGGUGGAUAAACUGCUGGAUGGAC--GUCUAUCUGUCUAUCUGGCUGCGAAUG -..............(((......)))((..(((((((--(......))))))))..))........ ( -17.00, z-score = -2.99, R) >droSec1.super_10 2913797 60 - 3036183 -GUGACUGACAAACUGGCGGAUAAACUGCUGGAUGG------ACGCUUGUCUAUGUGGCUGCGAGUG -((.((.......(..((((.....))))..)((((------((....)))))))).))........ ( -16.00, z-score = -1.85, R) >droSim1.chrX 2294123 60 - 17042790 -GUGACUGACAAACUGGCGGAUAAACUGCUGGAUGG------ACGCUUGUCUAUCUGGCUGCGAGUG -...........(((.(((......).((..(((((------((....)))))))..)).)).))). ( -20.30, z-score = -3.83, R) >droGri2.scaffold_14853 4390204 63 - 10151454 AUUGAUUGGCAGUCUGAUUGAUUCACAGAUUGACUG----AAUUGACGAUUGAGUGGCUUUUGAGUG (((.(..(((((((((.........)))))).(((.----((((...)))).))).)))..).))). ( -10.60, z-score = 0.23, R) >anoGam1.chr2R 42725602 63 + 62725911 -CUCAUUGUUACACACACACACACAC-GCCUAACCGC--ACGACGCAUAUCCGUCCGAGGUUGAUUU -.........................-...(((((..--..((((......))))...))))).... ( -8.50, z-score = -0.28, R) >consensus _GUGACUGACAAACUGGCGGAUAAACUGCUGGAUGG______ACGCCUGUCUAUCUGGCUGCGAGUG .............(((((((.....)))))))...........(((..(((.....))).))).... ( -4.44 = -4.39 + -0.05)
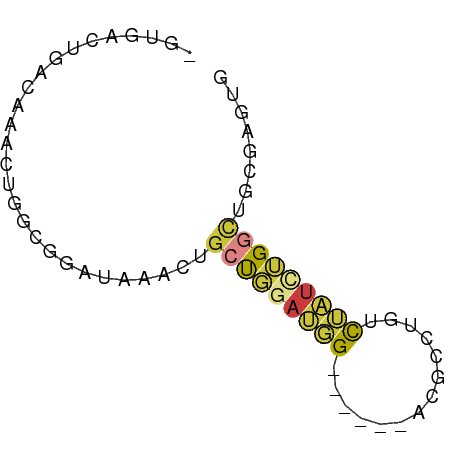
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:11:13 2011