| Sequence ID | dm3.chrX |
|---|---|
| Location | 3,205,474 – 3,205,618 |
| Length | 144 |
| Max. P | 0.546203 |

| Location | 3,205,474 – 3,205,618 |
|---|---|
| Length | 144 |
| Sequences | 7 |
| Columns | 154 |
| Reading direction | reverse |
| Mean pairwise identity | 66.90 |
| Shannon entropy | 0.65718 |
| G+C content | 0.55413 |
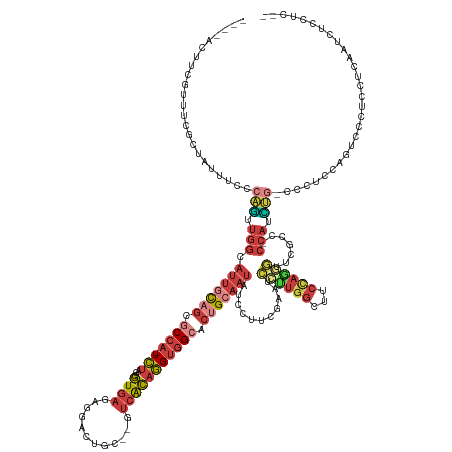
| Mean single sequence MFE | -40.75 |
| Consensus MFE | -14.34 |
| Energy contribution | -16.14 |
| Covariance contribution | 1.80 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.546203 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
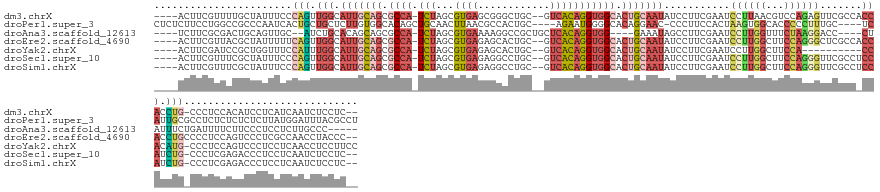
>dm3.chrX 3205474 144 - 22422827 ----ACUUCGUUUUGCUAUUUCCCAGUUGGCAUUGCAGCGCCA-UCUAGCGUGAGCGGGCUGC--GUCACAGGUGGCACUGCAAUAUCCUUCGAAUCCUUAACGUCCAGAGUUCGCCACCACCUG-CCCUCCACAUCCUCAUCAAUCUCCUC-- ----...................(((.(((((((((((.((((-(((.((....)).(((...--)))..))))))).))))))).......(((..((........))..)))))))....)))-..........................-- ( -36.80, z-score = -1.11, R) >droPer1.super_3 2849722 145 + 7375914 CUCUCUUCCUGGCCGCCCAAUCACUGCUGCUCUUGUGGCACAGCUGCAACUUAACGCCACUGC----AGAAUGGGGCACAGGAAC-CCCUUCCACUAAGUGGCACCCCCUUUGC----UCAUUGCGCCUCUCUCUCUCUUAUGGAUUUACGCCU .....((((((...((((........((((....(((((................))))).))----))....)))).)))))).-....((((.((((.((.........(((----.....)))........)).))))))))......... ( -35.72, z-score = 0.32, R) >droAna3.scaffold_12613 333090 134 + 519072 ----UCUUCGCGACUGCAGUUGC--AUCUGCACAGCAGCGCCA-UCUAGCGUGAAAAGGCCGCUGCUCACAGGUGG----GAAAUAGCCUUCGAAUCCUUGGUUUCUAAGGACC----CUAUUUCUGAUUUUCUUCCCUCCUCUUGCCC----- ----.....(((((....)))))--....(((.(((((((((.-((......))...)).)))))))...(((.((----(((((((((((.(((.(....).))).))))...----))))))).((......)))).)))..)))..----- ( -38.50, z-score = -0.69, R) >droEre2.scaffold_4690 616255 145 - 18748788 ----ACUUCGUUACGCUAUUUUUCAGUUGGCAUUGCAGCGCCA-UCUAGCGUGAGAGCACUGC--GUCACAGGUGGCACUGCAAUAUCCUUCGAAUCCUUGGCUUCCAGGGCUCGCCACCACCUGCCCCUCCAGUCCCUCGCCAACCUACCC-- ----.....................(((((((((((((.((((-(((...((((..((...))--.))))))))))).)))))))...............((((....((((............))))....))))....))))))......-- ( -45.00, z-score = -2.22, R) >droYak2.chrX 16593292 136 + 21770863 ----ACUUCGAUCCGCUGGUUUCCAUUUGGCAUUGCAGCGCCA-UCUAGCGUGAGAGCACUGC--GUCACAGGUGGCACUGCAAUAUCCUUCGAAUCCUUGGCUUCCA----------CCACAUG-CCCUCCAGUCCCUCCUCAACCUCCUUCC ----.....((...(((((....(((.(((.(((((((.((((-(((...((((..((...))--.))))))))))).))))))).......(((.(....).)))..----------))).)))-....)))))...)).............. ( -37.80, z-score = -1.83, R) >droSec1.super_10 2912264 144 - 3036183 ----ACUUCGUUUCGCUAUUUCCCAGUUGGCAUUGCAGCGCCA-UCUAGCGUGAGAGGCCUGC--GUCACAGGUGGCACUGCAAUAUCCUUCGAAUCCUUGGCUUCCAGGGUUCGCCUCCAUCUG-CCCUCGAGACCCUCCUCAAUCUCCUC-- ----.....((((((........(((..((((((((((.((((-(((.((((.((....))))--))...))))))).))))))).......(((((((.((...)))))))))))).....)))-....))))))................-- ( -45.70, z-score = -2.02, R) >droSim1.chrX 2292602 144 - 17042790 ----ACUUCGUUUCGCUAUUUCCCAGUUGGCAUUGCAGCGCCA-UCUAGCGUGAGAGGCCUGC--GUCACAGGUGGCACUGCAAUAUCCUUCGAAUCCUUGGCUUCCAGGGUUCGCCUCCAUCUG-CCCUCGAGACCCUCCUCAAUCUCCUC-- ----.....((((((........(((..((((((((((.((((-(((.((((.((....))))--))...))))))).))))))).......(((((((.((...)))))))))))).....)))-....))))))................-- ( -45.70, z-score = -2.02, R) >consensus ____ACUUCGUUUCGCUAUUUCCCAGUUGGCAUUGCAGCGCCA_UCUAGCGUGAGAGGACUGC__GUCACAGGUGGCACUGCAAUAUCCUUCGAAUCCUUGGCUUCCAGGGUUCGCC_CCAUCUG_CCCUCCAGUCCCUCCUCAAUCUCCUC__ .......................(((.(((.(((((((.((((.(((...((((............))))))))))).)))))))...........((((((...)))))).......))).)))............................. (-14.34 = -16.14 + 1.80)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:11:12 2011