| Sequence ID | dm3.chrX |
|---|---|
| Location | 3,194,809 – 3,194,976 |
| Length | 167 |
| Max. P | 0.586700 |

| Location | 3,194,809 – 3,194,976 |
|---|---|
| Length | 167 |
| Sequences | 5 |
| Columns | 182 |
| Reading direction | reverse |
| Mean pairwise identity | 71.11 |
| Shannon entropy | 0.47755 |
| G+C content | 0.35641 |
| Mean single sequence MFE | -31.17 |
| Consensus MFE | -12.26 |
| Energy contribution | -14.26 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.586700 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
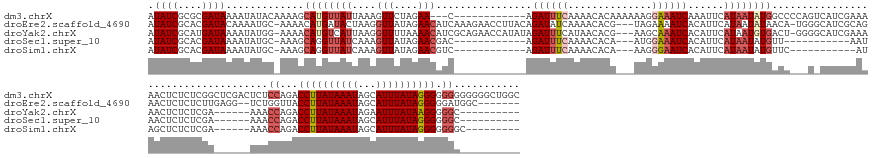
>dm3.chrX 3194809 167 - 22422827 AUAUCGCGCGAUAAAAUAUACAAAAGCAUGUUAUUAAAGUUCUAGAA---C------------AGAUUUCAAAACACAAAAAAGGAAAUCAAAUUCAUAAUAUGGCCCCAGUCAUCGAAAAACUCUCUCGGCUCGACUCUCCAGACCUUAUAAAUAGCAUUUAUAGGGGGGGGGGGGCUGGC .((((....)))).............((((((((....(((....))---)------------.((((((..............))))))......))))))))(((((((((.((((.........))))...))))((((...((((((((((...)))))))))).))))))))).... ( -41.64, z-score = -2.66, R) >droEre2.scaffold_4690 604506 168 - 18748788 AUAUCGCACGAUACAAAAUGC-AAAACAUGAUACUAAGGUUAUAGAACAUCAAAGAACCUUACAGAUAUCAAAACACG---UAGAAAAUCACAUUCAUAAUAUAACA-UGGGCAUCGCAGAACUCUCUCUUGAGG--UCUGGUUACCUUAUAAAUAGCAUUUAUAGGGGGAUGGC------- .((((....))))......((-......(((((((((((((...(.....)....))))))..)).))))).......---..........(((((......((((.-(((((.(((.(((......)))))).)--))))))))((((((((((...)))))))))))))))))------- ( -31.50, z-score = -1.29, R) >droYak2.chrX 16582452 161 + 21770863 AUAUCGCAUGAUAAAAUAUGG-AAAACAUGUCAUUAAGGUUUUAAAACAUCGCAGAACCAUAUAGAUUUCAUAACACG---AAGCAAAUCACAUUCAUAAUGUGACU-GGGGCAUCGAAAAACUCUCUCGA------AAACCAGACCUUAUAAAUAGAAUUUAUAAGGGGGC---------- ...(((.((((....(((((.-....)))))......((((((..........)))))).........))))....))---).((...((((((.....))))))((-((....((((.........))))------...)))).((((((((((...))))))))))..))---------- ( -30.80, z-score = -1.61, R) >droSec1.super_10 2901283 139 - 3036183 AUAUCGCACGAUAAAAUAUGC-AAAGCAGGUUAUCAAAGUUAUAGAACGAC------------AGAUUUCAAAACACA---AUGGAAAUCACAUUCAUAAUAUGUU-----------AAUAACUCUCUCGA------AAACCAGACCUUAUAAAUAGCAUUUAUAGGGGGGC---------- ...(((...(((((....(((-...)))..)))))..((((((........------------.((((((........---...))))))((((.......)))).-----------.))))))....)))------...((...((((((((((...)))))))))).)).---------- ( -24.70, z-score = -2.03, R) >droSim1.chrX 2282338 140 - 17042790 AUAUCGCACGAUAAAAUAUGC-AAAGCAGGUUAUCAAAGUUAUAGAACGUC------------AGAUUUCAAAACACA---AAGGGAAUCACAUUCAUAAUAUGUUC-----------AUAGCUCUCUCGA------AAACCAGACCUUAUAAAUAGCAUUUAUAGGGGGGGC--------- ...(((...(((((....(((-...)))..)))))..((((((.((((((.------------.((((((........---...))))))...........))))))-----------))))))....)))------...((...((((((((((...)))))))))).))..--------- ( -27.22, z-score = -2.39, R) >consensus AUAUCGCACGAUAAAAUAUGC_AAAGCAUGUUAUUAAAGUUAUAGAACAUC____________AGAUUUCAAAACACA___AAGGAAAUCACAUUCAUAAUAUGACC___G_CAUCGAAAAACUCUCUCGA______AAACCAGACCUUAUAAAUAGCAUUUAUAGGGGGGC__________ .((((....)))).............((((((((....(((....)))................((((((..............))))))......))))))))....................................((...((((((((((...)))))))))).))........... (-12.26 = -14.26 + 2.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:11:11 2011