
| Sequence ID | dm3.chrX |
|---|---|
| Location | 3,190,133 – 3,190,227 |
| Length | 94 |
| Max. P | 0.872971 |

| Location | 3,190,133 – 3,190,227 |
|---|---|
| Length | 94 |
| Sequences | 7 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 66.95 |
| Shannon entropy | 0.59555 |
| G+C content | 0.49129 |
| Mean single sequence MFE | -28.47 |
| Consensus MFE | -9.92 |
| Energy contribution | -12.37 |
| Covariance contribution | 2.45 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.608736 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
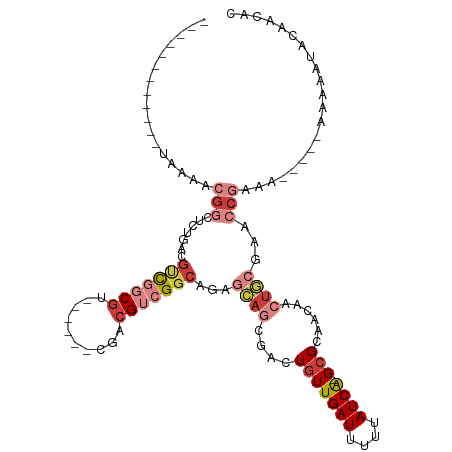
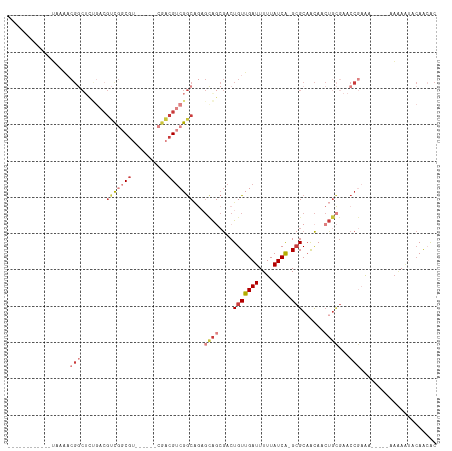
>dm3.chrX 3190133 94 + 22422827 ------------UAAAACGGCUCUGACGUCUGCGU------CGACGUCGGCAGAGCAGCGACUGUUGAUUUUUAUCA-GCGCAACAACUGCGAACCGAAA-----AAAAAUACAAAAC ------------.....(((((((((((((.....------.)))))))).)).((((....(((((((....))))-)))......))))...)))...-----............. ( -27.30, z-score = -2.03, R) >droSim1.chrX 2277834 94 + 17042790 ------------UAAAACGGCUCUGACGUCGGCGU------CGACGUCGGCAGAGCAGCGACUGUUGAUUUUUAUCA-GCGCAACAACUGCGAACCGAAA-----AAAAGUACAGCAC ------------.......((((((((((((....------)))))))...))))).((.(((((((((....))))-))(((.....))).........-----...)))...)).. ( -29.90, z-score = -1.56, R) >droSec1.super_10 2896770 95 + 3036183 ------------UAAAACGGCUCUGACGUCGGCGU------CGACGUCGGCAGAGCAGCGACUGUUGAUUUUUAUCA-GCGCAACAACUGCGAACCGAAA----AAAAAGUUCAACAC ------------.....((((((((((((((....------))))))))).)).((((....(((((((....))))-)))......))))...)))...----.............. ( -29.30, z-score = -1.63, R) >droYak2.chrX 16577711 99 - 21770863 ------------UAAAACGGCUCUGACGUCGGCGUAGCCGUCGACGUCGGCCGAGCAGCGACUGUUGAUUUUUAUCA-GCGCAACAACUGCGAACCGAAA------AGAACACAACAC ------------.....(((((..((((((((((....))))))))))))))).((((....(((((((....))))-)))......)))).........------............ ( -35.90, z-score = -2.81, R) >droEre2.scaffold_4690 599976 93 + 18748788 ------------UAAAACGGC------GUCGCCGU------CGACGUCGGCAGCGCAGCGACUGUUGAUUUUUAUCA-GCGCAACAACUGCGAACCGAAAGAAAAAAAAAUACAACAC ------------.....((((------((((....------))))))))....(((((....(((((((....))))-)))......)))))...(....)................. ( -25.90, z-score = -1.37, R) >droAna3.scaffold_12613 313453 110 - 519072 CGAAACGGCGCCAACGCCGACUGCGCCGCUGACGGCGACUGCAGCGCCGGCGUUGUGCCGACUGUUGAUUUUUAUCA-GAGCAGCAAC-AAAAACCACAA------UAACAACAACAC .....((((((.(((((((.((((((((....))))....))))...))))))))))))).((((((((....))))-..))))....-...........------............ ( -38.00, z-score = -2.45, R) >droGri2.scaffold_14853 4366081 81 + 10151454 ------------------------CACAACAGCAAC-----AAAAGUCGACACACAA--AACUGUUGAUUUUUAUCGCGCGCCGUAAAUGUGAAAAACGU------AUAAUGAAAAGC ------------------------.......((...-----(((((((((((.....--...)))))))))))...))((..(((..((((.....))))------...)))....)) ( -13.00, z-score = 0.01, R) >consensus ____________UAAAACGGCUCUGACGUCGGCGU______CGACGUCGGCAGAGCAGCGACUGUUGAUUUUUAUCA_GCGCAACAACUGCGAACCGAAA_____AAAAAUACAACAC .................(((.......((((((((........))))))))...((((....(((((((....))))...)))....))))...)))..................... ( -9.92 = -12.37 + 2.45)

| Location | 3,190,133 – 3,190,227 |
|---|---|
| Length | 94 |
| Sequences | 7 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 66.95 |
| Shannon entropy | 0.59555 |
| G+C content | 0.49129 |
| Mean single sequence MFE | -32.37 |
| Consensus MFE | -12.62 |
| Energy contribution | -14.54 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.872971 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
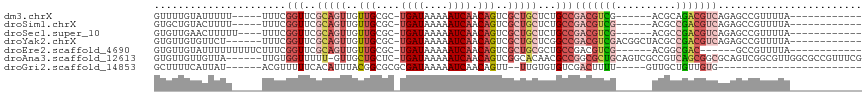
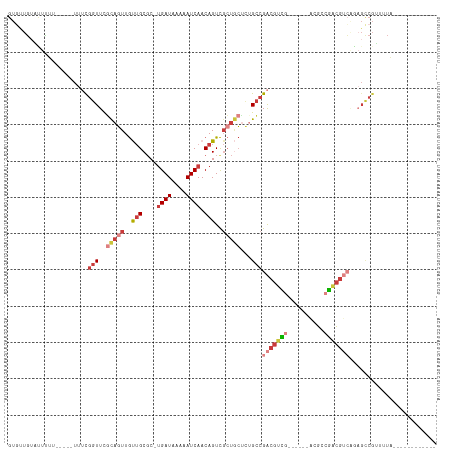
>dm3.chrX 3190133 94 - 22422827 GUUUUGUAUUUUU-----UUUCGGUUCGCAGUUGUUGCGC-UGAUAAAAAUCAACAGUCGCUGCUCUGCCGACGUCG------ACGCAGACGUCAGAGCCGUUUUA------------ .............-----...((((((((((..((.((((-((...........))).))).)).)))).((((((.------.....)))))).)))))).....------------ ( -32.60, z-score = -2.82, R) >droSim1.chrX 2277834 94 - 17042790 GUGCUGUACUUUU-----UUUCGGUUCGCAGUUGUUGCGC-UGAUAAAAAUCAACAGUCGCUGCUCUGCCGACGUCG------ACGCCGACGUCAGAGCCGUUUUA------------ .............-----...((((((((((..((.((((-((...........))).))).)).)))).(((((((------....))))))).)))))).....------------ ( -34.10, z-score = -2.56, R) >droSec1.super_10 2896770 95 - 3036183 GUGUUGAACUUUUU----UUUCGGUUCGCAGUUGUUGCGC-UGAUAAAAAUCAACAGUCGCUGCUCUGCCGACGUCG------ACGCCGACGUCAGAGCCGUUUUA------------ ..............----...((((((((((..((.((((-((...........))).))).)).)))).(((((((------....))))))).)))))).....------------ ( -34.10, z-score = -2.61, R) >droYak2.chrX 16577711 99 + 21770863 GUGUUGUGUUCU------UUUCGGUUCGCAGUUGUUGCGC-UGAUAAAAAUCAACAGUCGCUGCUCGGCCGACGUCGACGGCUACGCCGACGUCAGAGCCGUUUUA------------ ............------...(((((((((((..(((...-((((....)))).)))..)))))......(((((((.((....)).))))))).)))))).....------------ ( -34.10, z-score = -1.03, R) >droEre2.scaffold_4690 599976 93 - 18748788 GUGUUGUAUUUUUUUUUCUUUCGGUUCGCAGUUGUUGCGC-UGAUAAAAAUCAACAGUCGCUGCGCUGCCGACGUCG------ACGGCGAC------GCCGUUUUA------------ (((((((.............(((((.((((((..(((...-((((....)))).)))..))))))..)))))((...------.)))))))------)).......------------ ( -28.30, z-score = -0.83, R) >droAna3.scaffold_12613 313453 110 + 519072 GUGUUGUUGUUA------UUGUGGUUUUU-GUUGCUGCUC-UGAUAAAAAUCAACAGUCGGCACAACGCCGGCGCUGCAGUCGCCGUCAGCGGCGCAGUCGGCGUUGGCGCCGUUUCG .(((((((((((------..(..((....-...))..)..-))))))....)))))..((((.((((((((..(((((.(((((.....))))))))))))))))))..))))..... ( -46.70, z-score = -2.66, R) >droGri2.scaffold_14853 4366081 81 - 10151454 GCUUUUCAUUAU------ACGUUUUUCACAUUUACGGCGCGCGAUAAAAAUCAACAGUU--UUGUGUGUCGACUUUU-----GUUGCUGUUGUG------------------------ ............------..............(((((((.(((((((((.((.(((...--.....))).)).))))-----))))))))))))------------------------ ( -16.70, z-score = -0.37, R) >consensus GUGUUGUAUUUUU_____UUUCGGUUCGCAGUUGUUGCGC_UGAUAAAAAUCAACAGUCGCUGCUCUGCCGACGUCG______ACGCCGACGUCAGAGCCGUUUUA____________ ......................(((..(((((..(((....((((....)))).)))..)))))...)))(((((((..........)))))))........................ (-12.62 = -14.54 + 1.92)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:11:09 2011