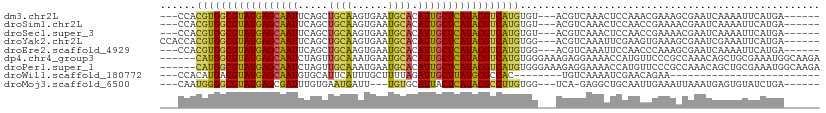
| Sequence ID | dm3.chr2L |
|---|---|
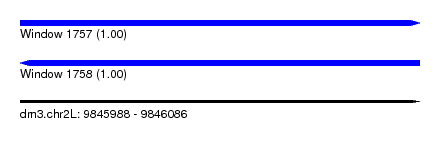
| Location | 9,845,988 – 9,846,086 |
| Length | 98 |
| Max. P | 0.999946 |

| Location | 9,845,988 – 9,846,086 |
|---|---|
| Length | 98 |
| Sequences | 9 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 65.17 |
| Shannon entropy | 0.70195 |
| G+C content | 0.44046 |
| Mean single sequence MFE | -31.29 |
| Consensus MFE | -17.70 |
| Energy contribution | -18.43 |
| Covariance contribution | 0.73 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.09 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.94 |
| SVM RNA-class probability | 0.999926 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
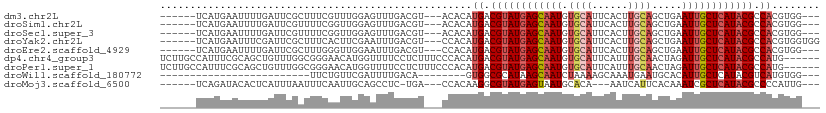
>dm3.chr2L 9845988 98 + 23011544 ------UCAUGAAUUUUGAUUCGCUUUCGUUUGGAGUUUGACGU---ACACAUGACGUAUGAGCAAUGUGCAUUCACUUGCAGCUGAAUUGCUCAUACGCCACGUGG--- ------....((((((..(..((....)).)..)))))).....---.(((.((.((((((((((((.((((......)))).....)))))))))))).)).))).--- ( -30.90, z-score = -2.18, R) >droSim1.chr2L 9627358 98 + 22036055 ------UCAUGAAUUUUGAUUCGUUUUCGGUUGGAGUUUGACGU---ACACAUGACGUAUGAGCAAUGUGCAUUCACUUGCAGCUGAAUUGCUCAUACGCCACGUGG--- ------....((((((..((.((....))))..)))))).....---.(((.((.((((((((((((.((((......)))).....)))))))))))).)).))).--- ( -33.00, z-score = -2.71, R) >droSec1.super_3 5292666 98 + 7220098 ------UCAUGAAUUUUGAUUCGUUUUCGGUUGGAGUUUGACGU---ACACAUGACGUAUGAGCAAUGUGCAUUCACUUGCAGCUGAAUUGCUCAUACGCCACGUGG--- ------....((((((..((.((....))))..)))))).....---.(((.((.((((((((((((.((((......)))).....)))))))))))).)).))).--- ( -33.00, z-score = -2.71, R) >droYak2.chr2L 12534910 101 + 22324452 ------UCAUGAAUUUCGAUUCGCUUUCACUUCGAAUUUGACGU---CCACAUGACGUAUGAGCAAUGUGCAUUCACUUGCAGCUGAAUUGCUCAUACGCCACGUGGUGG ------(((.....(((((............)))))..)))((.---((((.((.((((((((((((.((((......)))).....)))))))))))).)).)))))). ( -33.50, z-score = -3.00, R) >droEre2.scaffold_4929 10458112 98 + 26641161 ------UCAUGAAUUUUGAUUCGCUUUGGGUUGGAAUUUGACGU---CCACAUGACGUAUGAGCAAUGUGCAUUCACUUGCAGCUGAAUUGCUCAUACGCCACGUGG--- ------....((((((..((((.....))))..)))))).....---((((.((.((((((((((((.((((......)))).....)))))))))))).)).))))--- ( -36.10, z-score = -3.74, R) >dp4.chr4_group3 11367668 104 - 11692001 UCUUGCCAUUUCGCAGCUGUUUGGCGGGAACAUGGUUUUCCUCUUUCCCACAUGACGUAUGAGCAAUGUGCAUUCAUUUGCAACUAGAUUGCUCAUACGCCAUG------ ....((((....((....)).))))(((((...((....))...))))).((((.((((((((((((.((((......)))).....)))))))))))).))))------ ( -39.80, z-score = -5.08, R) >droPer1.super_1 8483257 104 - 10282868 UCUUGCCAUUUCGCAGCUGUUUGGCGGGAACAUGGUUUUCCUCUUUCCCACAUGACGUAUGAGCAAUGUGCAUUCAUUUGCAACUAGAUUGCUCAUACGCCAUG------ ....((((....((....)).))))(((((...((....))...))))).((((.((((((((((((.((((......)))).....)))))))))))).))))------ ( -39.80, z-score = -5.08, R) >droWil1.scaffold_180772 8510244 74 + 8906247 -------------------------UUCUGUUCGAUUUUGACA--------GUGGCGCAUAAGCAAUCUAAAAGCAAAUGAAUGCACAUUGCUCAUACGUCAUGUGG--- -------------------------..(((((.......))))--------)(((((.((.((((((......(((......)))..)))))).)).))))).....--- ( -14.60, z-score = 0.26, R) >droMoj3.scaffold_6500 3664317 94 - 32352404 ------UCAGAUACACUCAUUUAAUUUCAAUUGCAGCCUC-UGA---CCACAAGGCGUAUGAGUAAUGCACA---AAUCAUUCACAAAUCGCUCAUACGCCCCAUUG--- ------(((((....((((.((......)).)).))..))-)))---......((((((((((..((.....---............))..))))))))))......--- ( -20.93, z-score = -3.56, R) >consensus ______UCAUGAAUUUUGAUUCGCUUUCAGUUGGAGUUUGACGU___CCACAUGACGUAUGAGCAAUGUGCAUUCACUUGCAGCUGAAUUGCUCAUACGCCACGUGG___ ....................................................((.((((((((((((.((((......)))).....)))))))))))).))........ (-17.70 = -18.43 + 0.73)
| Location | 9,845,988 – 9,846,086 |
|---|---|
| Length | 98 |
| Sequences | 9 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 65.17 |
| Shannon entropy | 0.70195 |
| G+C content | 0.44046 |
| Mean single sequence MFE | -34.88 |
| Consensus MFE | -25.34 |
| Energy contribution | -24.96 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.48 |
| Mean z-score | -4.18 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 5.10 |
| SVM RNA-class probability | 0.999946 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
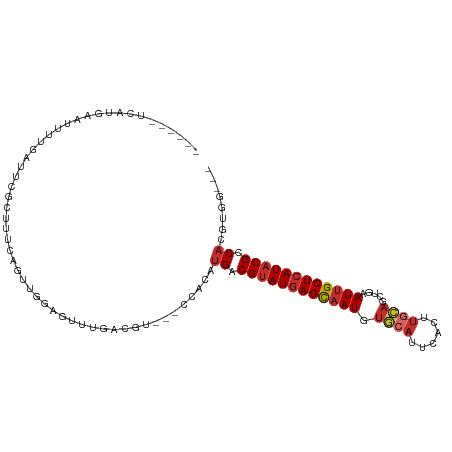
>dm3.chr2L 9845988 98 - 23011544 ---CCACGUGGCGUAUGAGCAAUUCAGCUGCAAGUGAAUGCACAUUGCUCAUACGUCAUGUGU---ACGUCAAACUCCAAACGAAAGCGAAUCAAAAUUCAUGA------ ---.(((((((((((((((((((...(.((((......)))))))))))))))))))))))).---...(((.........(....).((((....)))).)))------ ( -34.10, z-score = -4.02, R) >droSim1.chr2L 9627358 98 - 22036055 ---CCACGUGGCGUAUGAGCAAUUCAGCUGCAAGUGAAUGCACAUUGCUCAUACGUCAUGUGU---ACGUCAAACUCCAACCGAAAACGAAUCAAAAUUCAUGA------ ---.(((((((((((((((((((...(.((((......)))))))))))))))))))))))).---......................((((....))))....------ ( -33.40, z-score = -4.20, R) >droSec1.super_3 5292666 98 - 7220098 ---CCACGUGGCGUAUGAGCAAUUCAGCUGCAAGUGAAUGCACAUUGCUCAUACGUCAUGUGU---ACGUCAAACUCCAACCGAAAACGAAUCAAAAUUCAUGA------ ---.(((((((((((((((((((...(.((((......)))))))))))))))))))))))).---......................((((....))))....------ ( -33.40, z-score = -4.20, R) >droYak2.chr2L 12534910 101 - 22324452 CCACCACGUGGCGUAUGAGCAAUUCAGCUGCAAGUGAAUGCACAUUGCUCAUACGUCAUGUGG---ACGUCAAAUUCGAAGUGAAAGCGAAUCGAAAUUCAUGA------ ...((((((((((((((((((((...(.((((......)))))))))))))))))))))))))---.(((.((.(((((.((....))...))))).)).))).------ ( -43.80, z-score = -5.73, R) >droEre2.scaffold_4929 10458112 98 - 26641161 ---CCACGUGGCGUAUGAGCAAUUCAGCUGCAAGUGAAUGCACAUUGCUCAUACGUCAUGUGG---ACGUCAAAUUCCAACCCAAAGCGAAUCAAAAUUCAUGA------ ---((((((((((((((((((((...(.((((......)))))))))))))))))))))))))---......................((((....))))....------ ( -36.90, z-score = -4.91, R) >dp4.chr4_group3 11367668 104 + 11692001 ------CAUGGCGUAUGAGCAAUCUAGUUGCAAAUGAAUGCACAUUGCUCAUACGUCAUGUGGGAAAGAGGAAAACCAUGUUCCCGCCAAACAGCUGCGAAAUGGCAAGA ------(((((((((((((((((...((.(((......)))))))))))))))))))))).(((((...((....))...)))))((((.............)))).... ( -41.32, z-score = -4.79, R) >droPer1.super_1 8483257 104 + 10282868 ------CAUGGCGUAUGAGCAAUCUAGUUGCAAAUGAAUGCACAUUGCUCAUACGUCAUGUGGGAAAGAGGAAAACCAUGUUCCCGCCAAACAGCUGCGAAAUGGCAAGA ------(((((((((((((((((...((.(((......)))))))))))))))))))))).(((((...((....))...)))))((((.............)))).... ( -41.32, z-score = -4.79, R) >droWil1.scaffold_180772 8510244 74 - 8906247 ---CCACAUGACGUAUGAGCAAUGUGCAUUCAUUUGCUUUUAGAUUGCUUAUGCGCCAC--------UGUCAAAAUCGAACAGAA------------------------- ---.....((.((((((((((((.((((......)))....).)))))))))))).))(--------((((......).))))..------------------------- ( -21.10, z-score = -2.60, R) >droMoj3.scaffold_6500 3664317 94 + 32352404 ---CAAUGGGGCGUAUGAGCGAUUUGUGAAUGAUU---UGUGCAUUACUCAUACGCCUUGUGG---UCA-GAGGCUGCAAUUGAAAUUAAAUGAGUGUAUCUGA------ ---..(..(((((((((((..((.((..(.....)---..)).))..)))))))))))..)..---(((-((.((..((.(((....))).))...)).)))))------ ( -28.60, z-score = -2.40, R) >consensus ___CCACGUGGCGUAUGAGCAAUUCAGCUGCAAGUGAAUGCACAUUGCUCAUACGUCAUGUGG___ACGUCAAACUCCAACCGAAAGCGAAUCAAAAUUCAUGA______ ......(((((((((((((((((.....((((......)))).))))))))))))))))).................................................. (-25.34 = -24.96 + -0.39)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:29:05 2011