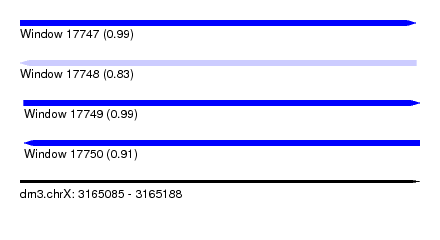
| Sequence ID | dm3.chrX |
|---|---|
| Location | 3,165,085 – 3,165,188 |
| Length | 103 |
| Max. P | 0.990543 |

| Location | 3,165,085 – 3,165,187 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 95.63 |
| Shannon entropy | 0.07566 |
| G+C content | 0.39051 |
| Mean single sequence MFE | -27.72 |
| Consensus MFE | -27.14 |
| Energy contribution | -27.34 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.98 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.22 |
| SVM RNA-class probability | 0.985983 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
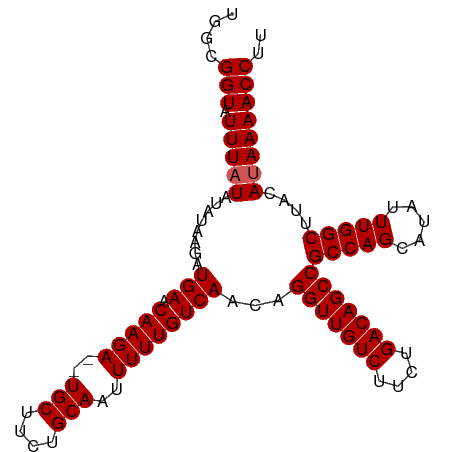
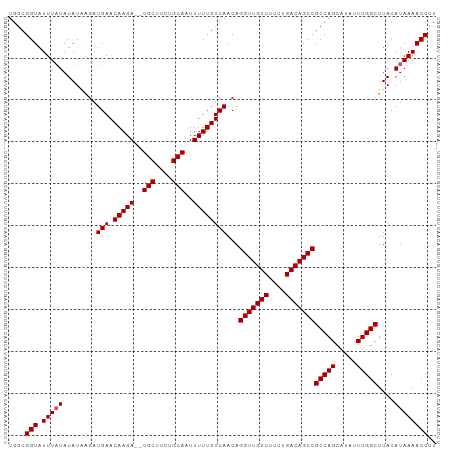
>dm3.chrX 3165085 102 + 22422827 UGACGGUAUUUUUAUAUAAGAUGAACAAGAGAUGCUUCUGCAAUUUUUGUCAACAGGUUGUCUUCUGACAGCCGCCAGCAUAUUUGGCUUACAUAAAACCUU ....(((...(((((.((((.(((.((((((.(((....))).))))))))).).(((((((....)))))))(((((.....)))))))).)))))))).. ( -29.20, z-score = -3.35, R) >droSim1.chrX 2253691 100 + 17042790 UGGCGGUAUUUAUAUAUAAGAUGAACAAGA--UGCUCCUGCAAUUUUUGUCAACAGGUUGUCUUCUGACAGCCGCCAGCAUAUUUGGCUUACAUAAAACCUU ....(((.(((((......(.(((.(((((--(((....)))..)))))))).).(((((((....)))))))(((((.....)))))....)))))))).. ( -27.40, z-score = -2.39, R) >droSec1.super_10 2872644 100 + 3036183 UGGCGGUAUUUAUAUAUAAGAUGAACAAGA--UGCUCCUGCAAUUUUUGUCAACAGGUUGUCUUCUGACAGCCGCCAGCAUAUUUGGCUUACAUAAAACCUU ....(((.(((((......(.(((.(((((--(((....)))..)))))))).).(((((((....)))))))(((((.....)))))....)))))))).. ( -27.40, z-score = -2.39, R) >droYak2.chrX 16552424 100 - 21770863 UGGCGGUGUUUAUAUAUAAAAUGAACAAGA--UGCUUCUGCAGUUUUUGUCAACAGGUUGUCUUCUGACAGCCGCCAGCAUAUUUGGCUUACAUAAAACCUU .((((.(((((((.......)))))))...--))))......((((((((.....(((((((....)))))))(((((.....)))))..))).)))))... ( -27.40, z-score = -1.99, R) >droEre2.scaffold_4690 573806 100 + 18748788 UGGCGGUAUUUAUAUAUGGGAUGAACAAGA--UGCUUCUGCAGUUUUUGUCAACAGGUUGUCUUCUGACAGCCGCCAGCAUAUUUGGCUUACAUAAAACCUU ....(((.(((((........(((.(((((--(((....)))..))))))))...(((((((....)))))))(((((.....)))))....)))))))).. ( -27.20, z-score = -1.41, R) >consensus UGGCGGUAUUUAUAUAUAAGAUGAACAAGA__UGCUUCUGCAAUUUUUGUCAACAGGUUGUCUUCUGACAGCCGCCAGCAUAUUUGGCUUACAUAAAACCUU ....(((.(((((........(((.(((((..(((....)))..))))))))...(((((((....)))))))(((((.....)))))....)))))))).. (-27.14 = -27.34 + 0.20)

| Location | 3,165,085 – 3,165,187 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 95.63 |
| Shannon entropy | 0.07566 |
| G+C content | 0.39051 |
| Mean single sequence MFE | -23.86 |
| Consensus MFE | -23.62 |
| Energy contribution | -23.82 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.99 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.828747 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 3165085 102 - 22422827 AAGGUUUUAUGUAAGCCAAAUAUGCUGGCGGCUGUCAGAAGACAACCUGUUGACAAAAAUUGCAGAAGCAUCUCUUGUUCAUCUUAUAUAAAAAUACCGUCA ..((((((((((((((((.......))))((.((((....)))).))....(((((....(((....)))....)))))....)))))))))...))).... ( -23.70, z-score = -1.56, R) >droSim1.chrX 2253691 100 - 17042790 AAGGUUUUAUGUAAGCCAAAUAUGCUGGCGGCUGUCAGAAGACAACCUGUUGACAAAAAUUGCAGGAGCA--UCUUGUUCAUCUUAUAUAUAAAUACCGCCA ..(((((((((((.((((.......)))).((((((....)))..(((((...........)))))))).--..............)))))))).))).... ( -24.90, z-score = -1.32, R) >droSec1.super_10 2872644 100 - 3036183 AAGGUUUUAUGUAAGCCAAAUAUGCUGGCGGCUGUCAGAAGACAACCUGUUGACAAAAAUUGCAGGAGCA--UCUUGUUCAUCUUAUAUAUAAAUACCGCCA ..(((((((((((.((((.......)))).((((((....)))..(((((...........)))))))).--..............)))))))).))).... ( -24.90, z-score = -1.32, R) >droYak2.chrX 16552424 100 + 21770863 AAGGUUUUAUGUAAGCCAAAUAUGCUGGCGGCUGUCAGAAGACAACCUGUUGACAAAAACUGCAGAAGCA--UCUUGUUCAUUUUAUAUAUAAACACCGCCA ..(((((((((((.((((.......))))((.((((....)))).))....(((((....(((....)))--..))))).......)))))))).))).... ( -22.90, z-score = -0.94, R) >droEre2.scaffold_4690 573806 100 - 18748788 AAGGUUUUAUGUAAGCCAAAUAUGCUGGCGGCUGUCAGAAGACAACCUGUUGACAAAAACUGCAGAAGCA--UCUUGUUCAUCCCAUAUAUAAAUACCGCCA ..(((((((((((.((((.......))))((.((((....)))).))....(((((....(((....)))--..))))).......)))))))).))).... ( -22.90, z-score = -1.03, R) >consensus AAGGUUUUAUGUAAGCCAAAUAUGCUGGCGGCUGUCAGAAGACAACCUGUUGACAAAAAUUGCAGAAGCA__UCUUGUUCAUCUUAUAUAUAAAUACCGCCA ..(((((((((((.((((.......))))((.((((....)))).))....(((((....(((....)))....))))).......)))))))).))).... (-23.62 = -23.82 + 0.20)

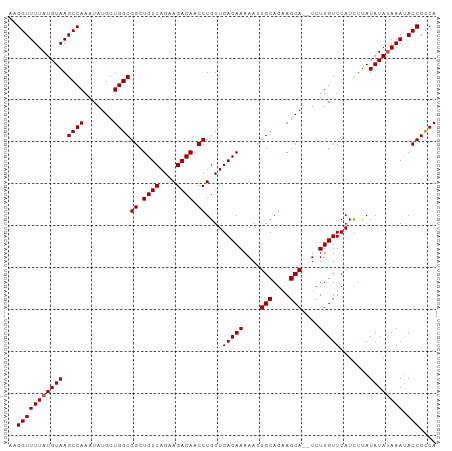
| Location | 3,165,086 – 3,165,188 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 95.63 |
| Shannon entropy | 0.07566 |
| G+C content | 0.39051 |
| Mean single sequence MFE | -27.68 |
| Consensus MFE | -27.14 |
| Energy contribution | -27.34 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.98 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.43 |
| SVM RNA-class probability | 0.990543 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
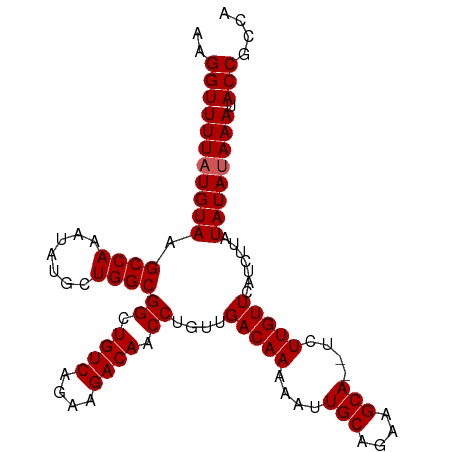
>dm3.chrX 3165086 102 + 22422827 GACGGUAUUUUUAUAUAAGAUGAACAAGAGAUGCUUCUGCAAUUUUUGUCAACAGGUUGUCUUCUGACAGCCGCCAGCAUAUUUGGCUUACAUAAAACCUUU ...(((...(((((.((((.(((.((((((.(((....))).))))))))).).(((((((....)))))))(((((.....)))))))).))))))))... ( -29.20, z-score = -3.54, R) >droSim1.chrX 2253692 100 + 17042790 GGCGGUAUUUAUAUAUAAGAUGAACAAGA--UGCUCCUGCAAUUUUUGUCAACAGGUUGUCUUCUGACAGCCGCCAGCAUAUUUGGCUUACAUAAAACCUUU ...(((.(((((......(.(((.(((((--(((....)))..)))))))).).(((((((....)))))))(((((.....)))))....))))))))... ( -27.40, z-score = -2.58, R) >droSec1.super_10 2872645 100 + 3036183 GGCGGUAUUUAUAUAUAAGAUGAACAAGA--UGCUCCUGCAAUUUUUGUCAACAGGUUGUCUUCUGACAGCCGCCAGCAUAUUUGGCUUACAUAAAACCUUU ...(((.(((((......(.(((.(((((--(((....)))..)))))))).).(((((((....)))))))(((((.....)))))....))))))))... ( -27.40, z-score = -2.58, R) >droYak2.chrX 16552425 100 - 21770863 GGCGGUGUUUAUAUAUAAAAUGAACAAGA--UGCUUCUGCAGUUUUUGUCAACAGGUUGUCUUCUGACAGCCGCCAGCAUAUUUGGCUUACAUAAAACCUUU ((((.(((((((.......)))))))...--))))......((((((((.....(((((((....)))))))(((((.....)))))..))).))))).... ( -27.20, z-score = -2.11, R) >droEre2.scaffold_4690 573807 100 + 18748788 GGCGGUAUUUAUAUAUGGGAUGAACAAGA--UGCUUCUGCAGUUUUUGUCAACAGGUUGUCUUCUGACAGCCGCCAGCAUAUUUGGCUUACAUAAAACCUUU ...(((.(((((........(((.(((((--(((....)))..))))))))...(((((((....)))))))(((((.....)))))....))))))))... ( -27.20, z-score = -1.61, R) >consensus GGCGGUAUUUAUAUAUAAGAUGAACAAGA__UGCUUCUGCAAUUUUUGUCAACAGGUUGUCUUCUGACAGCCGCCAGCAUAUUUGGCUUACAUAAAACCUUU ...(((.(((((........(((.(((((..(((....)))..))))))))...(((((((....)))))))(((((.....)))))....))))))))... (-27.14 = -27.34 + 0.20)



| Location | 3,165,086 – 3,165,188 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 95.63 |
| Shannon entropy | 0.07566 |
| G+C content | 0.39051 |
| Mean single sequence MFE | -23.86 |
| Consensus MFE | -23.62 |
| Energy contribution | -23.82 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.99 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.908428 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 3165086 102 - 22422827 AAAGGUUUUAUGUAAGCCAAAUAUGCUGGCGGCUGUCAGAAGACAACCUGUUGACAAAAAUUGCAGAAGCAUCUCUUGUUCAUCUUAUAUAAAAAUACCGUC ...((((((((((((((((.......))))((.((((....)))).))....(((((....(((....)))....)))))....)))))))))...)))... ( -23.70, z-score = -1.77, R) >droSim1.chrX 2253692 100 - 17042790 AAAGGUUUUAUGUAAGCCAAAUAUGCUGGCGGCUGUCAGAAGACAACCUGUUGACAAAAAUUGCAGGAGCA--UCUUGUUCAUCUUAUAUAUAAAUACCGCC ...(((((((((((.((((.......)))).((((((....)))..(((((...........)))))))).--..............)))))))).)))... ( -24.90, z-score = -1.56, R) >droSec1.super_10 2872645 100 - 3036183 AAAGGUUUUAUGUAAGCCAAAUAUGCUGGCGGCUGUCAGAAGACAACCUGUUGACAAAAAUUGCAGGAGCA--UCUUGUUCAUCUUAUAUAUAAAUACCGCC ...(((((((((((.((((.......)))).((((((....)))..(((((...........)))))))).--..............)))))))).)))... ( -24.90, z-score = -1.56, R) >droYak2.chrX 16552425 100 + 21770863 AAAGGUUUUAUGUAAGCCAAAUAUGCUGGCGGCUGUCAGAAGACAACCUGUUGACAAAAACUGCAGAAGCA--UCUUGUUCAUUUUAUAUAUAAACACCGCC ...(((((((((((.((((.......))))((.((((....)))).))....(((((....(((....)))--..))))).......)))))))).)))... ( -22.90, z-score = -1.15, R) >droEre2.scaffold_4690 573807 100 - 18748788 AAAGGUUUUAUGUAAGCCAAAUAUGCUGGCGGCUGUCAGAAGACAACCUGUUGACAAAAACUGCAGAAGCA--UCUUGUUCAUCCCAUAUAUAAAUACCGCC ...(((((((((((.((((.......))))((.((((....)))).))....(((((....(((....)))--..))))).......)))))))).)))... ( -22.90, z-score = -1.29, R) >consensus AAAGGUUUUAUGUAAGCCAAAUAUGCUGGCGGCUGUCAGAAGACAACCUGUUGACAAAAAUUGCAGAAGCA__UCUUGUUCAUCUUAUAUAUAAAUACCGCC ...(((((((((((.((((.......))))((.((((....)))).))....(((((....(((....)))....))))).......)))))))).)))... (-23.62 = -23.82 + 0.20)


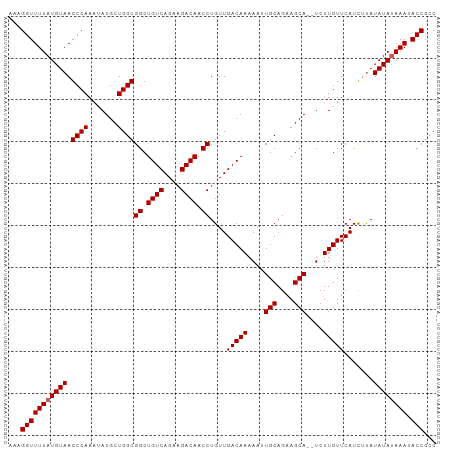
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:11:07 2011