| Sequence ID | dm3.chrX |
|---|---|
| Location | 3,106,391 – 3,106,496 |
| Length | 105 |
| Max. P | 0.637394 |

| Location | 3,106,391 – 3,106,496 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 76.30 |
| Shannon entropy | 0.42924 |
| G+C content | 0.41975 |
| Mean single sequence MFE | -26.15 |
| Consensus MFE | -15.43 |
| Energy contribution | -15.22 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.619775 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
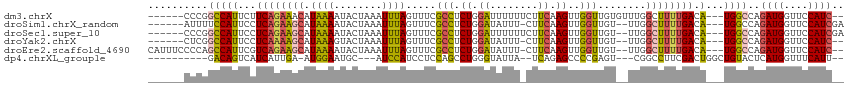
>dm3.chrX 3106391 105 + 22422827 ------CCCGGCCAUUCUUCAGAAACAUAAAAUACUAAAUUUAGUUUCGCCUCUGGAUUUUUUCUUCAAGUUGGUUGUGUUUGGCUUUUGACA---UGGCCAGAUGGUUCCAUC-- ------...((((((((...((.(((((((...(((((((((((........))))))))........)))...)))))))...))...)).)---))))).((((....))))-- ( -24.80, z-score = -1.14, R) >droSim1.chrX_random 1443549 104 + 5698898 ------AUUUUCCAUUCCUCAGAAGCAUAAAAUACUAAAUUUAGUUUCGCCUCUGGAUAUUU-CUUCAAGUUGGUUGU--UUGGCUUUUGACA---UGGCCAGAUGGUUCCAUCGA ------.....((((...((((((((...(((((((((((((((........))))))....-.......)))).)))--)).)))))))).)---)))...((((....)))).. ( -21.80, z-score = -0.54, R) >droSec1.super_10 2827088 105 + 3036183 ------CCCGGCCAUUCCUCAGAAGCAUAAAAUACUAAAUUUAGUUUCGCCUCUGGAUUUUUUCUUCAAGUUGGUUGU--UUGGCUUUUGACA---UGGCCAGAUGGUUCCAUCGA ------...((((((...((((((((...((((((((....))))...(((.((.((........)).))..))).))--)).)))))))).)---))))).((((....)))).. ( -30.00, z-score = -2.28, R) >droYak2.chrX 16501514 102 - 21770863 ------CUCGGCCAUUCCUCAAAAGCAUAAAGUACUAAAUUUAGUUUCGCCUCUGGAUAUUU-CUUCAAGUUGGUUGU--UUGGCUUUUGACA---UGGCCAGAUGGUUCCAUC-- ------...((((((...((((((((.((((..(((((((((((........))))))....-.......)))))..)--))))))))))).)---))))).((((....))))-- ( -27.90, z-score = -2.42, R) >droEre2.scaffold_4690 524398 108 + 18748788 CAUUUCCCCAGCCAUUCGUCAGAAGCAUAAAAUACUAAAUUUAGUUUCGCCUCUGGAUAUUU-CUUCAAGUUGGUUGU--UUGGCUUUUGACA---UGGCCAGAUGGUUCCAUC-- ..........(((((..(((((((((...(((((((((((((((........))))))....-.......)))).)))--)).))))))))))---))))..((((....))))-- ( -27.00, z-score = -1.92, R) >dp4.chrXL_group1e 2623799 95 + 12523060 ----------GACAGUCAUCAUUGA-AUGGAAUGC---AUCCAUCCUCCAGCCUGGGUAUUA--UCAGAGCCCCGAGU---CGGCCUUCGACUGGCUGUACUCAUGGUUUCAUU-- ----------..((.(((....)))-.)).((((.---..((((......((((((......--)))).))...((((---(((((.......))))).))))))))...))))-- ( -25.40, z-score = -0.02, R) >consensus ______CCCGGCCAUUCCUCAGAAGCAUAAAAUACUAAAUUUAGUUUCGCCUCUGGAUAUUU_CUUCAAGUUGGUUGU__UUGGCUUUUGACA___UGGCCAGAUGGUUCCAUC__ ..........((((....((((((((.((((........)))).....(((.((.((........)).))..)))........)))))))).....))))..((((....)))).. (-15.43 = -15.22 + -0.21)
| Location | 3,106,391 – 3,106,496 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 76.30 |
| Shannon entropy | 0.42924 |
| G+C content | 0.41975 |
| Mean single sequence MFE | -25.05 |
| Consensus MFE | -14.69 |
| Energy contribution | -15.04 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.637394 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
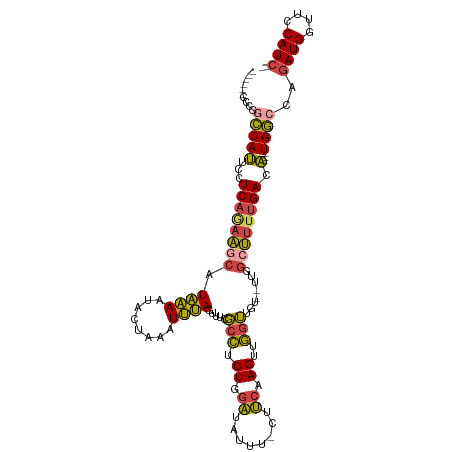
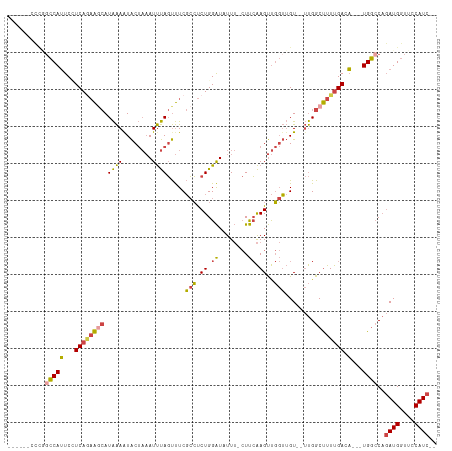
>dm3.chrX 3106391 105 - 22422827 --GAUGGAACCAUCUGGCCA---UGUCAAAAGCCAAACACAACCAACUUGAAGAAAAAAUCCAGAGGCGAAACUAAAUUUAGUAUUUUAUGUUUCUGAAGAAUGGCCGGG------ --((((....))))((((((---(.(((...(((.....(((.....)))..((.....))....)))(((((((((........)))).))))))))...)))))))..------ ( -25.50, z-score = -1.91, R) >droSim1.chrX_random 1443549 104 - 5698898 UCGAUGGAACCAUCUGGCCA---UGUCAAAAGCCAA--ACAACCAACUUGAAG-AAAUAUCCAGAGGCGAAACUAAAUUUAGUAUUUUAUGCUUCUGAGGAAUGGAAAAU------ ..((((....))))((((..---........)))).--....(((......(.-...).((((((((((..((((....))))......)))))))).))..))).....------ ( -19.40, z-score = -0.47, R) >droSec1.super_10 2827088 105 - 3036183 UCGAUGGAACCAUCUGGCCA---UGUCAAAAGCCAA--ACAACCAACUUGAAGAAAAAAUCCAGAGGCGAAACUAAAUUUAGUAUUUUAUGCUUCUGAGGAAUGGCCGGG------ ..((((....))))((((((---(..(.........--.(((.....)))...........((((((((..((((....))))......)))))))).)..)))))))..------ ( -25.90, z-score = -1.52, R) >droYak2.chrX 16501514 102 + 21770863 --GAUGGAACCAUCUGGCCA---UGUCAAAAGCCAA--ACAACCAACUUGAAG-AAAUAUCCAGAGGCGAAACUAAAUUUAGUACUUUAUGCUUUUGAGGAAUGGCCGAG------ --((((....))))((((((---(.((((((((...--........(((...(-(....))..))).....((((....)))).......))))))))...)))))))..------ ( -26.20, z-score = -2.50, R) >droEre2.scaffold_4690 524398 108 - 18748788 --GAUGGAACCAUCUGGCCA---UGUCAAAAGCCAA--ACAACCAACUUGAAG-AAAUAUCCAGAGGCGAAACUAAAUUUAGUAUUUUAUGCUUCUGACGAAUGGCUGGGGAAAUG --((((....))))....((---(.((...(((((.--.(((.....)))...-.......((((((((..((((....))))......)))))))).....)))))...)).))) ( -24.00, z-score = -1.01, R) >dp4.chrXL_group1e 2623799 95 - 12523060 --AAUGAAACCAUGAGUACAGCCAGUCGAAGGCCG---ACUCGGGGCUCUGA--UAAUACCCAGGCUGGAGGAUGGAU---GCAUUCCAU-UCAAUGAUGACUGUC---------- --........(((.(...((((((((((.....))---))).(((.......--.....))).)))))..(((((((.---....)))))-))..).)))......---------- ( -29.30, z-score = -1.06, R) >consensus __GAUGGAACCAUCUGGCCA___UGUCAAAAGCCAA__ACAACCAACUUGAAG_AAAUAUCCAGAGGCGAAACUAAAUUUAGUAUUUUAUGCUUCUGAGGAAUGGCCGGG______ ..((((....))))((((((.....((((..................))))..........((((((((....((((........)))))))))))).....))))))........ (-14.69 = -15.04 + 0.34)



Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:10:58 2011