| Sequence ID | dm3.chrX |
|---|---|
| Location | 3,064,654 – 3,064,757 |
| Length | 103 |
| Max. P | 0.819955 |

| Location | 3,064,654 – 3,064,757 |
|---|---|
| Length | 103 |
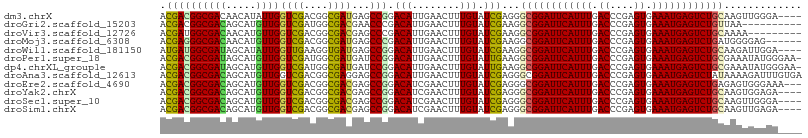
| Sequences | 12 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 87.34 |
| Shannon entropy | 0.27737 |
| G+C content | 0.50984 |
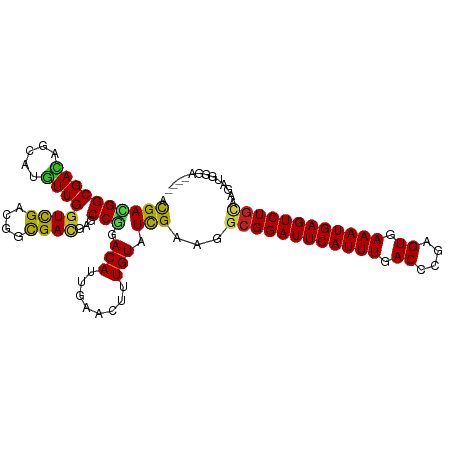
| Mean single sequence MFE | -33.42 |
| Consensus MFE | -26.48 |
| Energy contribution | -25.97 |
| Covariance contribution | -0.52 |
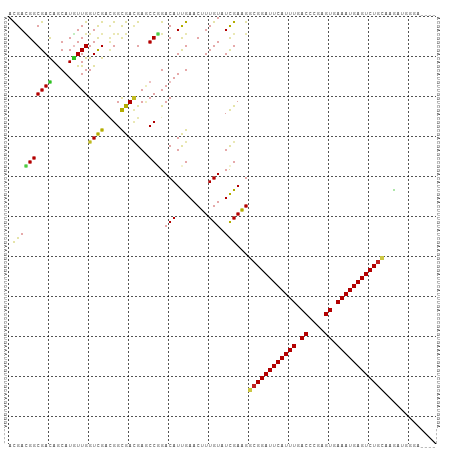
| Combinations/Pair | 1.32 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.819955 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 3064654 103 - 22422827 ACGACGGCGACAACAUAUUGGUCGACGGCGAUGAGCCGGACAUUGAACUUUGUAUCGAGGGCGGAUUCAUUUGACCCGAGUGAAAUGAGUCUGCAAGUUGGGA---- ..........((((.......((((((((.....)))).(((........))).))))..((((((((((((.((....)).))))))))))))..))))...---- ( -34.00, z-score = -1.86, R) >droGri2.scaffold_15203 2669664 97 - 11997470 ACGACGGCGACAGCAUGUUGGUCGAUGGCGACGAACCCGACAUUGAACUUUGUAUCGAAGGCGGAUUCAUUUGACCCGAGUGAAAUGAGUCUGUUAA---------- .(((((((....)).))))).((((((.((.......)).)))))).(((((...)))))((((((((((((.((....)).))))))))))))...---------- ( -30.40, z-score = -1.54, R) >droVir3.scaffold_12726 469108 98 - 2840439 ACGAUGGCGACAACAUGUUGGUCGACGGCGACGAGCCCGACAUUGAACUUUGUAUCGAAGGCGGAUUCAUUUGACCCGAGUGAAAUGAGUCUGCAAAA--------- .(((((.((((.....))))((((..(((.....)))))))...........)))))...((((((((((((.((....)).))))))))))))....--------- ( -34.60, z-score = -2.78, R) >droMoj3.scaffold_6308 2151634 101 - 3356042 ACGAGGGCGACAACAUGUUGGUCGACGGCGAUGAGCCCGACAUUGAACUUUGUAUCGAAGGCGGAUUCAUUUGACCCGAGUGAAAUGAGUCUGAUGGGGAG------ ((((((.((((.....))))((((..(((.....)))))))......)))))).((.....(((((((((((.((....)).))))))))))).....)).------ ( -31.70, z-score = -1.32, R) >droWil1.scaffold_181150 1692548 103 - 4952429 AUGAUGGCGAUAGCAUAUUGGUUGAAGGUGAUGAGCCGGACAUUGAACUUUGUAUCGAAGGCGGAUUCAUUUGACCCGAGUGAAAUGAGUCUGCAAGAUUGGA---- ..((((((....))...((((((.(......).)))))).))))...(((((...)))))((((((((((((.((....)).)))))))))))).........---- ( -28.20, z-score = -1.63, R) >droPer1.super_18 1468903 106 - 1952607 ACGACGGCGAUAGCAUGUUGGUCGAUGGCGAUGAUCCGGACAUUGAACUUUGUAUUGAAGGCGGAUUCAUUUGACCCGAGUGAAAUGAGUCUGCGAAAUAUGGGAA- .(((((((....)).))))).((((((.((......))..))))))..((..((((....((((((((((((.((....)).))))))))))))..))))..))..- ( -33.70, z-score = -2.26, R) >dp4.chrXL_group1e 2570737 106 - 12523060 ACGACGGCGAUAGCAUGUUGGUCGAUGGCGAUGAUCCGGACAUUGAACUUUGUAUUGAAGGCGGAUUCAUUUGACCCGAGUGAAAUGAGUCUGCGAAAUAUGGGAA- .(((((((....)).))))).((((((.((......))..))))))..((..((((....((((((((((((.((....)).))))))))))))..))))..))..- ( -33.70, z-score = -2.26, R) >droAna3.scaffold_12613 164532 107 + 519072 ACGACGGCGACAGCAUGUUGGUCGACGGCGAGGAGCCGGACAUUGAACUUUGUAUCGAGGGCGGAUUCAUUUGACCCGAGUGAAAUGAGUCUAUAAAAGAUUUGUGA .(((((((....)).))))).((((((((.....)))).(((........))).))))....((((((((((.((....)).))))))))))............... ( -30.10, z-score = -0.64, R) >droEre2.scaffold_4690 490863 104 - 18748788 ACGACGGCGACAGCAUGUUGGUCGACGGCGACGAGCCGGACAUCGAACUUUGUAUCGAGGGCGGAUUCAUUUGACCCGAGUGAAAUGAGUCUGAGAGUGGGAAA--- .(((((((....)).)))))(((..((((.....))))))).((((........))))...(((((((((((.((....)).)))))))))))...........--- ( -32.70, z-score = -0.47, R) >droYak2.chrX 16467195 103 + 21770863 ACGACGGCGACAGCAUGUUGGUCGACGGCGACGAGCCGGACAUCGAACUUUGUAUCGAGGGCGGAUUCAUUUGACCCGAGUGAAAUGAGUCUGCAAGUGGAGA---- .(((((((....)).)))))(((..((((.....))))))).((((........))))..((((((((((((.((....)).)))))))))))).........---- ( -36.50, z-score = -1.51, R) >droSec1.super_10 2794266 103 - 3036183 ACGACGGCGACAGCAUGUUGGUCGACGGCGACGAGCCGGACAUCGAACUUUGUAUCGAGGGCGGAUUCAUUUGACCCGAGUGAAAUGAGUCUGCAAGUUGGGA---- .(((((((....)).)))))((((....))))...((.(((.((((........))))..((((((((((((.((....)).))))))))))))..))).)).---- ( -38.10, z-score = -1.89, R) >droSim1.chrX 2169040 103 - 17042790 ACGACGGCGACAGCAUGUUGGUCGACGGCGACGAGCCGGACAUCGAACUUUGUAUCGAGGGCGGAUUCAUUUGACCCGAGUGAAAUGAGUCUGCAAGUUGAGA---- .(((((((....)).))))).((((((((.....)))...(.((((........)))).)((((((((((((.((....)).))))))))))))..)))))..---- ( -37.40, z-score = -1.90, R) >consensus ACGACGGCGACAGCAUGUUGGUCGACGGCGACGAGCCGGACAUUGAACUUUGUAUCGAAGGCGGAUUCAUUUGACCCGAGUGAAAUGAGUCUGCAAGAUGGGA____ .((((((((((.....))))((((....))))...))).(((........))).)))...((((((((((((.((....)).))))))))))))............. (-26.48 = -25.97 + -0.52)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:10:54 2011