
| Sequence ID | dm3.chrX |
|---|---|
| Location | 3,061,743 – 3,061,852 |
| Length | 109 |
| Max. P | 0.811024 |

| Location | 3,061,743 – 3,061,852 |
|---|---|
| Length | 109 |
| Sequences | 14 |
| Columns | 121 |
| Reading direction | forward |
| Mean pairwise identity | 63.31 |
| Shannon entropy | 0.79677 |
| G+C content | 0.57979 |
| Mean single sequence MFE | -38.22 |
| Consensus MFE | -13.04 |
| Energy contribution | -12.44 |
| Covariance contribution | -0.59 |
| Combinations/Pair | 1.86 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.811024 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
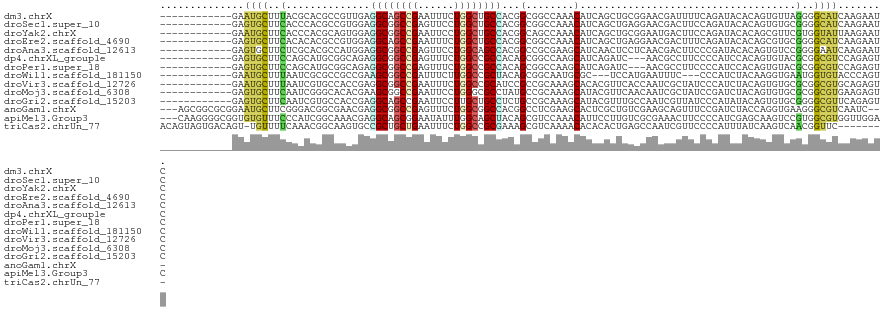
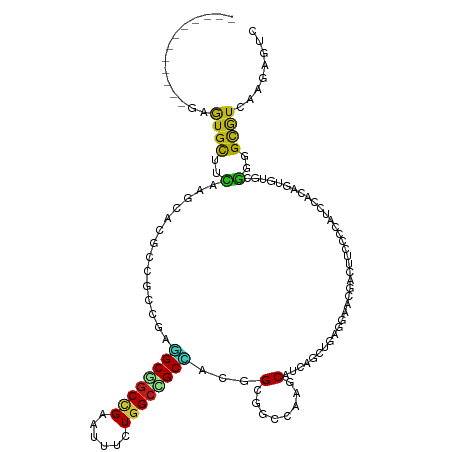
>dm3.chrX 3061743 109 + 22422827 ------------GAAUGCUUUACGCACGCCGUUGAGGCAGCCGAAUUUCUGGCUGCCACGGCGGCCAAACAUCAGCUGCGGAACGAUUUUCAGAUACACAGUGUUAGGGGCAUCAAGAAUC ------------((.((((((..(((((((((((.((((((((......)))))))).)))))))..........(((..((....))..))).......))))..))))))))....... ( -40.40, z-score = -3.02, R) >droSec1.super_10 2791391 109 + 3036183 ------------GAGUGCUUCACCCACGCCGUGGAGGCGGCCGAGUUCCUGGCUGCCACGGCGGCCAAACAUCAGCUGAGGAACGACUUCCAGAUACACAGUGUGCGGGGCAUCAAGAAUC ------------((.((((((.(.((((((((.(.((((((((......)))))))).).)))))..............((((....)))).........))).).))))))))....... ( -42.30, z-score = -1.24, R) >droYak2.chrX 16464284 109 - 21770863 ------------GAAUGCUUCACCCACGCAGUGGAGGCGGCCGAAUUCCUGGCUGCCACGGCAGCCAAACAUCAGCUGCGGAAUGACUUCCAGAUACACAGCGUUCGUGGUAUUAAGAAUC ------------.(((((..(((..((((.(((...(((((.((.....(((((((....)))))))....)).)))))((((....)))).....))).))))..))))))))....... ( -40.50, z-score = -2.64, R) >droEre2.scaffold_4690 488000 109 + 18748788 ------------GAGUGCUUCACACACGCCGUGGAGGCAGCCGAAUUUCUGGCUGCCACGGCGGCCAAACAUCAGCUGAGGAACGACUUUCAGAUACACAGCGUGCGGGGCAUCAAGAAUC ------------((.((((((.(((..(((((.(.((((((((......)))))))).).))))).........((((.(...(........)...).))))))).))))))))....... ( -45.30, z-score = -3.71, R) >droAna3.scaffold_12613 161071 109 - 519072 ------------GAGUGCUUCUCGCACGCCAUGGAGGCGGCCGAGUUCCUGGCAGCCACGGCCGCGAAGCAUCAACUCCUCAACGACUUCCCGAUACACAGUGUCCGGGGAAUCAAGAAUC ------------..((((.....)))).....(((((((((((.(((......)))..)))))))((....))..)))).....((.((((((((((...)))).)))))).))....... ( -38.30, z-score = -1.12, R) >dp4.chrXL_group1e 2567781 106 + 12523060 ------------GAGUGCUUCCAGCAUGCGGCAGAGGCGGCCGAGUUUCUGGCCGCCACAGCGGCCAAGCAUCAGAUC---AACGCCUUCCCCAUCCACAGUGUACGCGGCGUCCAGAGUC ------------((((((.....))))((.((.(.((((((((......)))))))).).)).))......)).....---.(((((.....(((.....))).....)))))........ ( -33.50, z-score = 0.39, R) >droPer1.super_18 1465934 106 + 1952607 ------------GAGUGCUUCCAGCAUGCGGCAGAGGCGGCCGAGUUUCUGGCCGCCACAGCGGCCAAGCAUCAGAUC---AACGCCUUCCCCAUCCACAGUGUACGCGGCGUCCAGAGUC ------------((((((.....))))((.((.(.((((((((......)))))))).).)).))......)).....---.(((((.....(((.....))).....)))))........ ( -33.50, z-score = 0.39, R) >droWil1.scaffold_181150 1689604 103 + 4952429 ------------GAAUGCUUUAAUCGCGCCGCCGAAGCGGCCGAUUUCUUGGCCGCUACAGCGGCAAUGCGC---UCCAUGAAUUUC---CCCAUCUACAAGGUGAAUGGUGUACCCAGUC ------------....((.......))(((((.(.(((((((((....))))))))).).))))).....((---.((((.....((---.((........)).)))))).))........ ( -34.80, z-score = -1.67, R) >droVir3.scaffold_12726 465950 109 + 2840439 ------------GAAUGCUUUAAUCGUGCCACCGAGGCGGCCGAAUUUCUGGCCGCAUCCGCCGCAAAGCACACGUUCACCAAUCGCUAUCCCAUCUACAGUGUGCGCGGCGUGCAGAGUC ------------....(((((......(((.....)))(((((......)))))(((..((((((...((((((..........................)))))))))))))))))))). ( -37.17, z-score = -1.27, R) >droMoj3.scaffold_6308 2147768 109 + 3356042 ------------GAGUGCUUCAAUCGGGCACACGAAGCGGCCGAAUUCCUGGGCGCCUAUGCCGCAAAGCAUACGUUCAACAAUCGCUAUCCGAUCUACAGUGUGCGCGGCGUGAAGAGUC ------------..((((((.....)))))).....(((.(((......))).))).((((((((...((((((........((((.....)))).....))))))))))))))....... ( -36.12, z-score = -0.38, R) >droGri2.scaffold_15203 2666340 109 + 11997470 ------------GAGUGCUUCAAUCGUGCCACCGAGGCAGCCGAAUUCCUUGCUGCCUCUGCCGCAAAGCAUACGUUUGCCAAUCGUUAUCCCAUAUACAGUGUGCGGGGCGUUCAGAGUC ------------(((((((((....(((.((..((((((((..........)))))))))).)))...((((((..........................)))))))))))))))...... ( -33.77, z-score = -0.94, R) >anoGam1.chrX 2645810 115 - 22145176 ---AGCGGCGCGGAAUGCUUCGGGACGGCGAACGAGGCGGCCGAGUUUCUGGCGGCCACGGCCUCGAAGCACUCGCUGUCGAAGCAGUUUCCGAUCUACCAGGUGAAGGGCGUCAAUC--- ---...(((((((((((((((..((((((((.(((((((((((.........)))))...))))))......))))))))))))))..))))......((.......)))))))....--- ( -51.10, z-score = -1.24, R) >apiMel3.Group3 2815279 118 + 12341916 ---CAAGGGGCGGUGUGUUUCCCAUCGGCAAACGAGGCAGCGGAAUAUUUGGCAGCUACAGCGUCCAAACAUUCCUUGUCGCGAAACUUCCCCAUCGAGCAAGUCCGUGGCGUGGUUGGAC ---...((((.(((..((((((....)).))))(.(((((.(((((.(((((..((....))..))))).)))))))))).)...))).)))).((((.((.((.....)).)).)))).. ( -37.80, z-score = 0.09, R) >triCas2.chrUn_77 196392 112 + 283703 ACAGUAGUGACAGU-UGUUUUCAAACGGCAAGUGCCGCUGCUGAAUUUCUGGCCGCGAAAGCGUCAAAACACACACUGAGCCAAUCGUUCCCCAUUUAUCAAGUCAACGGUUC-------- .((((((((.((.(-((((.......))))).)).)))))))).......((((((....)))(((..........))))))(((((((..(..........)..))))))).-------- ( -30.50, z-score = -1.39, R) >consensus ____________GAGUGCUUCAAGCACGCCGCCGAGGCGGCCGAAUUUCUGGCCGCCACGGCGGCCAAGCAUCAGCUGAGGAACGACUUCCCCAUCCACAGUGUGCGGGGCGUCAAGAGUC ..............((((..(..............((((((((......))))))))...(........)....................................)..))))........ (-13.04 = -12.44 + -0.59)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:10:53 2011