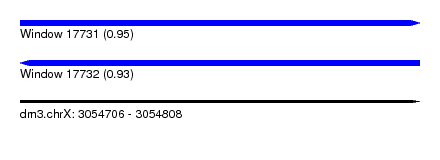
| Sequence ID | dm3.chrX |
|---|---|
| Location | 3,054,706 – 3,054,808 |
| Length | 102 |
| Max. P | 0.953423 |

| Location | 3,054,706 – 3,054,808 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 78.91 |
| Shannon entropy | 0.35714 |
| G+C content | 0.39475 |
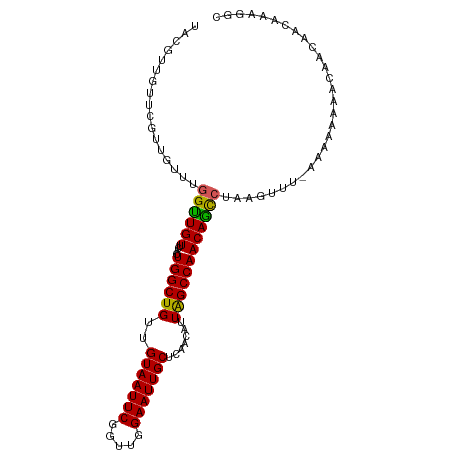
| Mean single sequence MFE | -25.01 |
| Consensus MFE | -18.84 |
| Energy contribution | -18.32 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.953423 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
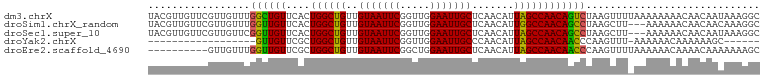
>dm3.chrX 3054706 102 + 22422827 UACGUUGUUCGUUGUUUGGCUGUUCACUGGCUGUUGUAAUUCGGUUGGAAUUGCUCAACAUUAGCCAACAGUCUAAGUUUUAAAAAAAACAACAAUAAAGGC ..(.(((((.(((((((((((((....((((((..(((((((.....))))))).......)))))))))))).............)))))))))))).).. ( -25.01, z-score = -1.64, R) >droSim1.chrX_random 1433301 99 + 5698898 UACGUUGUUCGUUGUUUGGUUGUUCACUGGCUGUUGUAAUUCGGUUGGAAUUGCUCAACAUUGGCCAACAGCCUAAGCUU---AAAAAACAACAACAAAGGC ..(.(((((.(((((((....(((....((((((((.......(((((......)))))......))))))))..)))..---...)))))))))))).).. ( -28.32, z-score = -1.81, R) >droSec1.super_10 2784447 99 + 3036183 UACGUUGUUCGUUGUUCGGUUGUUCACUGGCUGUUGUAAUUCGGUUGGAAUUGCUCAACAUUAGCCAACAGCCUAAGCUU---AAAAAACAACAAUAAAGGC ...((((((..(((...((((((....((((((..(((((((.....))))))).......))))))))))))......)---))..))))))......... ( -25.80, z-score = -1.69, R) >droYak2.chrX 16457226 77 - 21770863 ------------------GUUGUUCGCUGGCUGUUGUAAUUCGGUUGGAAUUGCCCAACAUUAGCCAACAACCCAAGUUU-AAAAAACAAAAAAGC------ ------------------((((((.(((((.(((((((((((.....)))))))..))))))))).))))))........-...............------ ( -19.30, z-score = -1.77, R) >droEre2.scaffold_4690 480965 92 + 18748788 ----------GUUGUUUGGUUGUUCGCUGGCUGUUGUAAUUCGGCUGGAAUUGCUCAACAUUAGCCAACAACCCAAGUUUUAAAAAACAAAACAAAAAAAGC ----------.(((((((((((((.(((((.(((((((((((.....)))))))..))))))))).)))))))...((((....)))).))))))....... ( -26.60, z-score = -2.73, R) >consensus UACGUUGUUCGUUGUUUGGUUGUUCACUGGCUGUUGUAAUUCGGUUGGAAUUGCUCAACAUUAGCCAACAGCCUAAGUUU_AAAAAAAACAACAACAAAGGC .................((((((....((((((..(((((((.....))))))).......))))))))))))............................. (-18.84 = -18.32 + -0.52)
| Location | 3,054,706 – 3,054,808 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 78.91 |
| Shannon entropy | 0.35714 |
| G+C content | 0.39475 |
| Mean single sequence MFE | -23.50 |
| Consensus MFE | -16.88 |
| Energy contribution | -17.48 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.932565 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 3054706 102 - 22422827 GCCUUUAUUGUUGUUUUUUUUAAAACUUAGACUGUUGGCUAAUGUUGAGCAAUUCCAACCGAAUUACAACAGCCAGUGAACAGCCAAACAACGAACAACGUA .......(((((((((..((((.....))))(((((.(((..(((((...(((((.....))))).)))))...))).)))))..)))))))))........ ( -21.00, z-score = -1.45, R) >droSim1.chrX_random 1433301 99 - 5698898 GCCUUUGUUGUUGUUUUUU---AAGCUUAGGCUGUUGGCCAAUGUUGAGCAAUUCCAACCGAAUUACAACAGCCAGUGAACAACCAAACAACGAACAACGUA ...(((((((((.......---..(((..((((((((((((....)).))(((((.....))))).))))))))))).........)))))))))....... ( -26.77, z-score = -2.15, R) >droSec1.super_10 2784447 99 - 3036183 GCCUUUAUUGUUGUUUUUU---AAGCUUAGGCUGUUGGCUAAUGUUGAGCAAUUCCAACCGAAUUACAACAGCCAGUGAACAACCGAACAACGAACAACGUA .......(((((((((...---.......(((((((((((.......)))(((((.....))))).)))))))).(....)....)))))))))........ ( -26.10, z-score = -2.47, R) >droYak2.chrX 16457226 77 + 21770863 ------GCUUUUUUGUUUUUU-AAACUUGGGUUGUUGGCUAAUGUUGGGCAAUUCCAACCGAAUUACAACAGCCAGCGAACAAC------------------ ------(((.....(((....-.)))...((((((((......((((((....)))))).......))))))))))).......------------------ ( -19.32, z-score = -1.36, R) >droEre2.scaffold_4690 480965 92 - 18748788 GCUUUUUUUGUUUUGUUUUUUAAAACUUGGGUUGUUGGCUAAUGUUGAGCAAUUCCAGCCGAAUUACAACAGCCAGCGAACAACCAAACAAC---------- .........((((((.....))))))...(((((((.(((..(((((...(((((.....))))).)))))...))).))))))).......---------- ( -24.30, z-score = -2.02, R) >consensus GCCUUUAUUGUUGUUUUUUUU_AAACUUAGGCUGUUGGCUAAUGUUGAGCAAUUCCAACCGAAUUACAACAGCCAGUGAACAACCAAACAACGAACAACGUA .............................(((((((.(((..(((((...(((((.....))))).)))))...))).)))))))................. (-16.88 = -17.48 + 0.60)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:10:52 2011