
| Sequence ID | dm3.chrX |
|---|---|
| Location | 3,047,653 – 3,047,745 |
| Length | 92 |
| Max. P | 0.922691 |

| Location | 3,047,653 – 3,047,745 |
|---|---|
| Length | 92 |
| Sequences | 8 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 65.06 |
| Shannon entropy | 0.68426 |
| G+C content | 0.44451 |
| Mean single sequence MFE | -18.21 |
| Consensus MFE | -7.43 |
| Energy contribution | -7.95 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.911605 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
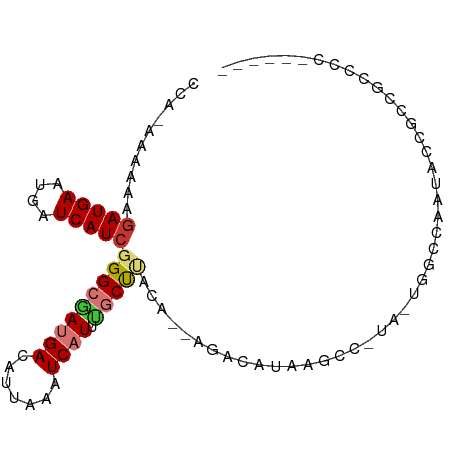
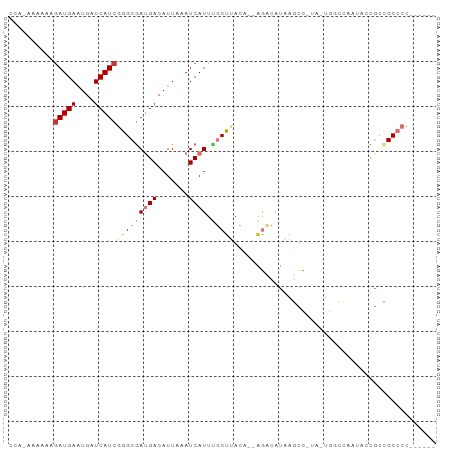
>dm3.chrX 3047653 92 + 22422827 CCAUAUAACAGAUGAAUGAUCAUCGGGCGAUGACAUUAAAUCAUUUGCUUACA--AGCCAUAAGUCCUA-UGGCCAAUAACAACGCCGCCCCCCC ..........(((((....)))))(((((((((.......))))(((.(((..--.((((((.....))-))))...))))))...))))).... ( -21.90, z-score = -2.59, R) >droSec1.super_10 2777573 88 + 3036183 CCAGAAAAAAAAUGAAUGAUCAUCGGGCGAUGACAUUAAAUCAUUUGCUUACA--AGCCAUGGGUC-UA-UGGCCAAUACCACCGCCGCCCC--- ...........((((....)))).(((((((((.......)))).........--.((((((....-))-))))............))))).--- ( -17.70, z-score = 0.01, R) >droYak2.chrX 16450017 81 - 21770863 ----AAAACCGAUGAAUGAUCAUCGGGCGAUGACAUUAAAUCAUUUGCCUACA--AGCCAUGAGUC-UA-UGGCCAAUACCGCCGCCCC------ ----......(((((....)))))(((((((((.......)))).........--.((((((....-))-)))).........))))).------ ( -21.60, z-score = -2.35, R) >droEre2.scaffold_4690 473895 85 + 18748788 UAAAAAAAAAGAUGAAUGAUCAUCGGGCGAUGACAUUAAAUCAUUUGCUUACA--AGCCAUGAGUC-UA-UGGCCAAUACCGCCGCCCC------ ..........(((((....)))))(((((((((.......)))).........--.((((((....-))-)))).........))))).------ ( -21.50, z-score = -2.63, R) >dp4.chrXL_group1e 2551785 80 + 12523060 -------AAAGAUGAAUGAUCAUCGGGCGAUGACAUUAAAUCAUACGCUCUCUC-AAACAUAAUCUAUA-UACACCCCACUGCAGCCCC------ -------...(((((....)))))(((((((........)))....((......-..............-...........)).)))).------ ( -13.98, z-score = -1.78, R) >droPer1.super_18 1449764 80 + 1952607 -------AAAGAUGAAUGAUCAUCGGGCGAUGACAUUAAAUCAUACGCUCUCUC-AAACAUAAUCUAU---ACACCCCACUGUAGCCCCCC---- -------...(((((....)))))(((((((........)))............-............(---(((......))))))))...---- ( -15.30, z-score = -2.33, R) >droVir3.scaffold_12726 449303 83 + 2840439 CCAGAGCAAAGAUGACUGAUCAUCGGGCAAAGACAUUAAAUCAUCCUCUGGUUAUGGAUACUAACUAUA---------ACUGCUGCACAUUC--- .(((.((...(((((....)))))..)).....................((((((((.......)))))---------))).))).......--- ( -15.60, z-score = -0.63, R) >droGri2.scaffold_15203 2648656 92 + 11997470 CCAGAGCAGAGAUGACUGAUCAUCGGGCAAAGACAUUAAAUCAUCUUCUGGUUAUGGAUACCGACUUACCUCUGUUGCACUGCUGCACAUUC--- ....(((((.(((((....)))))..(((((((...(((.((....(((......)))....)).)))..))).)))).)))))........--- ( -18.10, z-score = 0.59, R) >consensus CCA_AAAAAAGAUGAAUGAUCAUCGGGCGAUGACAUUAAAUCAUUUGCUUACA__AGACAUAAGCC_UA_UGGCCAAUACCGCCGCCCC______ ..........(((((....)))))(((((((((.......)))).)))))............................................. ( -7.43 = -7.95 + 0.52)

| Location | 3,047,653 – 3,047,745 |
|---|---|
| Length | 92 |
| Sequences | 8 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 65.06 |
| Shannon entropy | 0.68426 |
| G+C content | 0.44451 |
| Mean single sequence MFE | -24.62 |
| Consensus MFE | -10.70 |
| Energy contribution | -11.15 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.922691 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
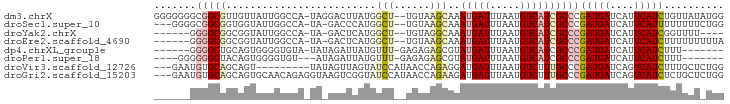
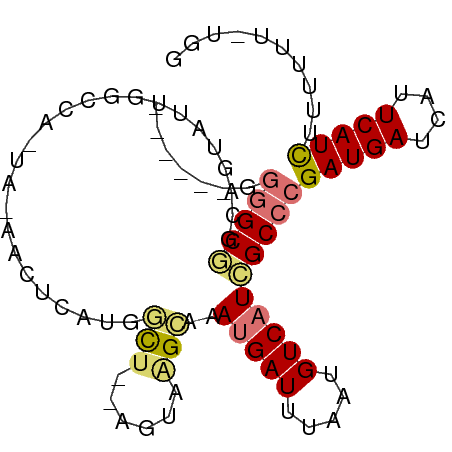
>dm3.chrX 3047653 92 - 22422827 GGGGGGGCGGCGUUGUUAUUGGCCA-UAGGACUUAUGGCU--UGUAAGCAAAUGAUUUAAUGUCAUCGCCCGAUGAUCAUUCAUCUGUUAUAUGG ....(((((...(((((((.(((((-(((...))))))))--.)).)))))(((((.....))))))))))(((((....))))).......... ( -29.50, z-score = -1.84, R) >droSec1.super_10 2777573 88 - 3036183 ---GGGGCGGCGGUGGUAUUGGCCA-UA-GACCCAUGGCU--UGUAAGCAAAUGAUUUAAUGUCAUCGCCCGAUGAUCAUUCAUUUUUUUUCUGG ---.(((((((.....(((.(((((-(.-.....))))))--.))).))..(((((.....))))))))))(((((....))))).......... ( -25.80, z-score = -0.81, R) >droYak2.chrX 16450017 81 + 21770863 ------GGGGCGGCGGUAUUGGCCA-UA-GACUCAUGGCU--UGUAGGCAAAUGAUUUAAUGUCAUCGCCCGAUGAUCAUUCAUCGGUUUU---- ------.(((((((..(((.(((((-(.-.....))))))--.))).))..(((((.....))))))))))(((((....)))))......---- ( -28.50, z-score = -2.38, R) >droEre2.scaffold_4690 473895 85 - 18748788 ------GGGGCGGCGGUAUUGGCCA-UA-GACUCAUGGCU--UGUAAGCAAAUGAUUUAAUGUCAUCGCCCGAUGAUCAUUCAUCUUUUUUUUUA ------.(((((((..(((.(((((-(.-.....))))))--.))).))..(((((.....))))))))))(((((....))))).......... ( -29.80, z-score = -3.59, R) >dp4.chrXL_group1e 2551785 80 - 12523060 ------GGGGCUGCAGUGGGGUGUA-UAUAGAUUAUGUUU-GAGAGAGCGUAUGAUUUAAUGUCAUCGCCCGAUGAUCAUUCAUCUUU------- ------.((((..(....)((((.(-((((((((((((((-....))))))..))))).))))))))))))(((((....)))))...------- ( -22.60, z-score = -1.28, R) >droPer1.super_18 1449764 80 - 1952607 ----GGGGGGCUACAGUGGGGUGU---AUAGAUUAUGUUU-GAGAGAGCGUAUGAUUUAAUGUCAUCGCCCGAUGAUCAUUCAUCUUU------- ----...((((....((((.((..---......(((((((-....))))))).......)).)))).))))(((((....)))))...------- ( -22.66, z-score = -1.38, R) >droVir3.scaffold_12726 449303 83 - 2840439 ---GAAUGUGCAGCAGU---------UAUAGUUAGUAUCCAUAACCAGAGGAUGAUUUAAUGUCUUUGCCCGAUGAUCAGUCAUCUUUGCUCUGG ---....(((((((...---------....))).))))......(((((((((((((.(.((((.......)))).).)))))))....)))))) ( -16.20, z-score = 0.19, R) >droGri2.scaffold_15203 2648656 92 - 11997470 ---GAAUGUGCAGCAGUGCAACAGAGGUAAGUCGGUAUCCAUAACCAGAAGAUGAUUUAAUGUCUUUGCCCGAUGAUCAGUCAUCUCUGCUCUGG ---...((..(....)..)).(((((.......(((.......)))(((.(((((((.(.((((.......)))).).)))))))))).))))). ( -21.90, z-score = -0.06, R) >consensus ______GGGGCGGCAGUAUUGGCCA_UA_AACUCAUGGCU__AGUAAGCAAAUGAUUUAAUGUCAUCGCCCGAUGAUCAUUCAUCUUUUUU_UGG .......(((((.........................((........))..(((((.....))))))))))(((((....))))).......... (-10.70 = -11.15 + 0.45)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:10:49 2011