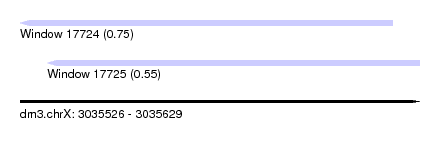
| Sequence ID | dm3.chrX |
|---|---|
| Location | 3,035,526 – 3,035,629 |
| Length | 103 |
| Max. P | 0.750768 |

| Location | 3,035,526 – 3,035,622 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 80.55 |
| Shannon entropy | 0.37278 |
| G+C content | 0.38391 |
| Mean single sequence MFE | -25.03 |
| Consensus MFE | -17.65 |
| Energy contribution | -16.85 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.750768 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 3035526 96 - 22422827 UGUCAGACGCUUGAAAUUUGAGAUACAUUUUCAAGUGGCAGCUUGAAGCAUGUGUUUAUCUCAGUGUUUGUGUAUCCUGCAGUUUC-AAAGGGGCCU ...((((((((.((......((((((((((((((((....)))))))).))))))))...))))))))))....((((........-..)))).... ( -25.70, z-score = -0.80, R) >droEre2.scaffold_4690 461628 97 - 18748788 UGUCAGACGCUUGAAAUUUGAGAUACAUUUUCAAGUGGCAGCUUGAAGCAUGUGUUUAUCGCAGUGUUUGUGUAUCCUGCAGUUUCAAAAGGUGUCU ....((((((((((((((..((((((((((((((((....)))))))).))))))))...((((..(......)..)))))))))))...))))))) ( -31.90, z-score = -3.14, R) >droYak2.chrX 16437545 96 + 21770863 UGUCAGACGCUUGAAAUUUGAGAUACAUUUUCAAGUGGCAUCUUGAAGCAUGUGUUUAUCUCAGUGUUUUUGUAUCCUGCCGUUUC-AAAGGUGUCU ....(((((((((((((...(((((((((((((((......))))))).))))))))....(((..(......)..)))..)))))-)..))))))) ( -25.20, z-score = -1.90, R) >droSec1.super_10 2765541 96 - 3036183 UGUCAGACGCUUGAAAUUUGAGAUACAUUUUCAAGUGGCAGCUUGAAGCAUGUGUUUAUCUCAGUGUUUGUGUAUCCUGCAGUUUC-AGAGGUGCCU ...((((((((.((......((((((((((((((((....)))))))).))))))))...)))))))))).((((((((......)-)).))))).. ( -29.30, z-score = -1.96, R) >droSim1.chrX 2159365 96 - 17042790 UGUCAGACGCUUGAAAUUCGAGAUACAUUUUCAAGUGGCAGCUUGAAGCAUGUGUUUAUCUCAGUGUUUGUGUAUCCUGCAGUUUC-AAAGGUGCCU ...((((((((.((......((((((((((((((((....)))))))).))))))))...)))))))))).(((.(((........-..)))))).. ( -26.30, z-score = -1.31, R) >triCas2.chrUn_35 58278 94 - 346786 UUUUAAACUUUUAACGUCU--UAUUCAGUUUCAGUUAGAAUUCUGAUACCUACGAAUUUUUUAGUUUUUAACAAUGACGUUUCUUC-AGAGAGAUUU .......(((((((((((.--......((.((((........)))).)).((.(((((....))))).)).....)))))).....-.))))).... ( -11.80, z-score = 0.40, R) >consensus UGUCAGACGCUUGAAAUUUGAGAUACAUUUUCAAGUGGCAGCUUGAAGCAUGUGUUUAUCUCAGUGUUUGUGUAUCCUGCAGUUUC_AAAGGUGCCU ...((((((((.((......(((((((((((((((......))))))).))))))))...))))))))))........(((.(((...))).))).. (-17.65 = -16.85 + -0.80)
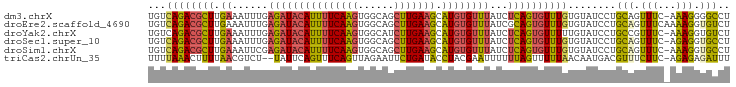
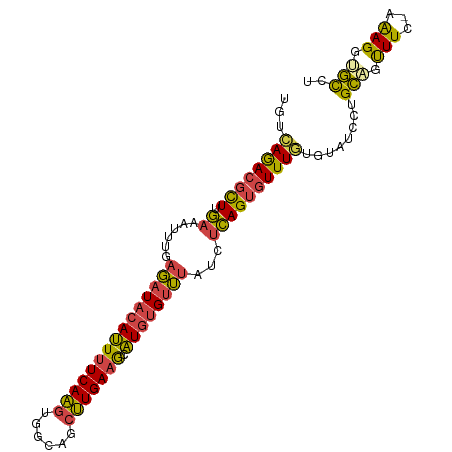
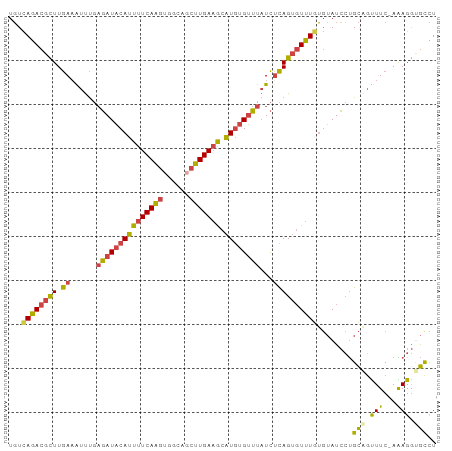
| Location | 3,035,533 – 3,035,629 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 74.73 |
| Shannon entropy | 0.43071 |
| G+C content | 0.39685 |
| Mean single sequence MFE | -26.75 |
| Consensus MFE | -15.51 |
| Energy contribution | -15.93 |
| Covariance contribution | 0.43 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.554133 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 3035533 96 - 22422827 -----------------CAGCGAGUGUCAGACGCUUGAAAUUUGAGAUACAUUUUCAAG-----UGGCAGCUUGAAGCAUGUGUUUAUCUCAGUGUUUGUGUAUCCUGCAGUUUCAAA- -----------------(((.((....((((((((.((......(((((((((((((((-----(....)))))))).))))))))...))))))))))....)))))..........- ( -27.90, z-score = -1.75, R) >droEre2.scaffold_4690 461635 97 - 18748788 -----------------CAGCGAGUGUCAGACGCUUGAAAUUUGAGAUACAUUUUCAAG-----UGGCAGCUUGAAGCAUGUGUUUAUCGCAGUGUUUGUGUAUCCUGCAGUUUCAAAA -----------------...((((((.....))))))...(((((((((((((((((((-----(....)))))))).)))).......((((..(......)..)))).)))))))). ( -27.90, z-score = -1.47, R) >droYak2.chrX 16437552 96 + 21770863 -----------------CAGCGAGUGUCAGACGCUUGAAAUUUGAGAUACAUUUUCAAG-----UGGCAUCUUGAAGCAUGUGUUUAUCUCAGUGUUUUUGUAUCCUGCCGUUUCAAA- -----------------(((.((.....(((((((.((......(((((((((((((((-----......))))))).))))))))...))))))))).....)))))..........- ( -22.60, z-score = -0.71, R) >droSec1.super_10 2765548 96 - 3036183 -----------------CAGCGAGUGUCAGACGCUUGAAAUUUGAGAUACAUUUUCAAG-----UGGCAGCUUGAAGCAUGUGUUUAUCUCAGUGUUUGUGUAUCCUGCAGUUUCAGA- -----------------(((.((....((((((((.((......(((((((((((((((-----(....)))))))).))))))))...))))))))))....)))))..........- ( -27.90, z-score = -1.45, R) >droSim1.chrX 2159372 96 - 17042790 -----------------CAGCGAGUGUCAGACGCUUGAAAUUCGAGAUACAUUUUCAAG-----UGGCAGCUUGAAGCAUGUGUUUAUCUCAGUGUUUGUGUAUCCUGCAGUUUCAAA- -----------------(((.((....((((((((.((......(((((((((((((((-----(....)))))))).))))))))...))))))))))....)))))..........- ( -27.90, z-score = -1.75, R) >dp4.chrXL_group1e 2537193 117 - 12523060 AUAUAAAUGUAUCUACGCUACUGGCAGUGUAUCUUUAACAUAGAAAGUGUAUCUUUCAGAUAUCUAUUAGCUUAGAA-AUGUAUCUACGCUACUGGCAGUGUAUCUUUAACAUUGCAG- ........(((((((((((......)))))............(((((.....)))))))))))....((((.((((.-.....)))).))))...(((((((.......)))))))..- ( -26.30, z-score = -0.98, R) >consensus _________________CAGCGAGUGUCAGACGCUUGAAAUUUGAGAUACAUUUUCAAG_____UGGCAGCUUGAAGCAUGUGUUUAUCUCAGUGUUUGUGUAUCCUGCAGUUUCAAA_ .................(((.((....((((((((.((......(((((((((((((((...........))))))).))))))))...))))))))))....)))))........... (-15.51 = -15.93 + 0.43)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:10:46 2011