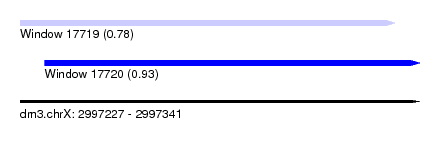
| Sequence ID | dm3.chrX |
|---|---|
| Location | 2,997,227 – 2,997,341 |
| Length | 114 |
| Max. P | 0.933403 |

| Location | 2,997,227 – 2,997,334 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 65.90 |
| Shannon entropy | 0.61462 |
| G+C content | 0.54828 |
| Mean single sequence MFE | -35.10 |
| Consensus MFE | -15.10 |
| Energy contribution | -15.68 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.779892 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
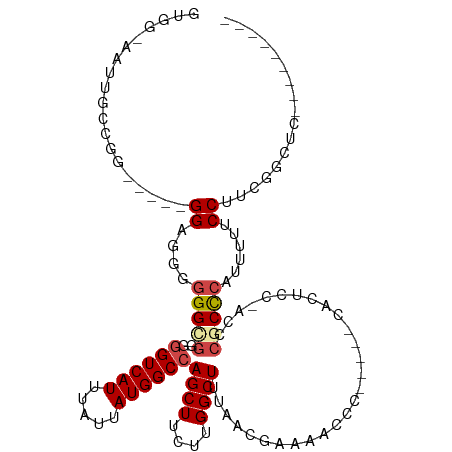
>dm3.chrX 2997227 107 + 22422827 GUGGCAAUUGCCGACG--GGGUGGGGGGUUGGGGUCAUUUAUUAUGGCCAGCUUCUUGGCUUCACCGAAAACCCACUACCACUCC-ACCCGCCCAUUUUUCCUACGGCUC--------- (((((....))).)).--(((((((((((..((((..........((((((....)))))).........))))...))).))))-))))(((............)))..--------- ( -41.31, z-score = -1.76, R) >droSim1.chrX 2122001 91 + 17042790 -------------------GGCGGGGGGCGGGGGUCAUUUAUUAUGGCCAGCUUCUUGGCUUUAACGAAAACCCACUACCACUCCCACCCGCCCAUUUUUCCUUCGGCUC--------- -------------------(((((((((.(((((((((.....))))))((((....)))).................)).).))).)))))).................--------- ( -32.70, z-score = -1.54, R) >droSec1.super_10 2727561 109 + 3036183 GUGGCAAUUGCCGGUGGCGGGCGGGGGGCGGGGGUCAUUUAUUAUGGCCAGCUUCUUGGCUUUAAAGAAAACCCACUACCACUCC-ACCCGCCCAUUUUUCCUUCUGCUC--------- .((((....))))(.((((((.((((((..(((((..(((.(((.((((((....)))))).))).))).)))).)..)).))))-.)))))))................--------- ( -45.20, z-score = -2.12, R) >droYak2.chrX 16397098 96 - 21770863 GUGGAAAUUGCCAGCA---GGGGGGAGGUGGAGGUCAUUUAUUAUGGCCAGCUUCUUGGCUUUUCCGAAACCC-------ACUCC-ACACGCCCAUUUCUCCCACUC------------ .(((......)))...---.(((((((((((.((((((.....))))))((((....))))............-------.....-......)))))))))))....------------ ( -31.10, z-score = -0.64, R) >droEre2.scaffold_4690 421116 100 + 18748788 GUGGAAAUUGU--------GGAGGAGGGGGUAGGUCAUUUAUUAUGGCCAGCUUCUUGGCUUUUCCGAAAAACC-----CACUCC-ACUCGCCCAUUUUUCCUUUGACUUGCUC----- ..(((((..((--------((..((((((((.(((..(((.....((((((....)))))).....)))..)))-----.)))))-.)))..)))).)))))............----- ( -32.30, z-score = -1.57, R) >droAna3.scaffold_12613 76259 104 - 519072 ----------CUGGGUCCUGGAGUUGGGGGCCAGUCAUUUAUUAUGGCCAGCUUCUUGGCUUUUACCUCCCCCC-----CUCCCCUACUCACUCCUCUAUCCACCCAACAAAGCCACUG ----------.(((((...(((((((((((...............((((((....)))))).............-----..))))))...))))).......)))))............ ( -28.01, z-score = -0.58, R) >consensus GUGG_AAUUGCCGG_____GGAGGGGGGCGGGGGUCAUUUAUUAUGGCCAGCUUCUUGGCUUUAACGAAAACCC_____CACUCC_ACCCGCCCAUUUUUCCUUCGGCUC_________ ...................((....(((((..((((((.....))))))((((....))))............................)))))......))................. (-15.10 = -15.68 + 0.58)

| Location | 2,997,234 – 2,997,341 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 67.33 |
| Shannon entropy | 0.56819 |
| G+C content | 0.54386 |
| Mean single sequence MFE | -32.67 |
| Consensus MFE | -19.52 |
| Energy contribution | -20.28 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.933403 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
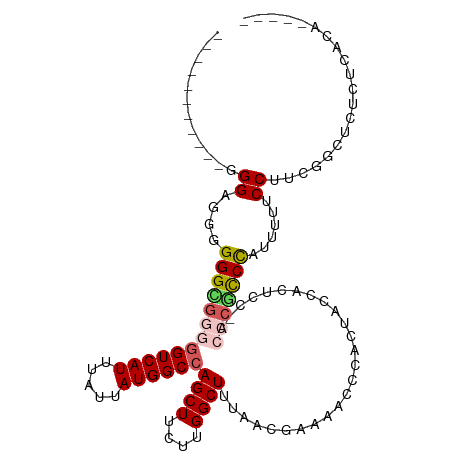
>dm3.chrX 2997234 107 + 22422827 UUGCCGACG--GGGUGGGGGGUUGGGGUCAUUUAUUAUGGCCAGCUUCUUGGCUUCACCGAAAACCCACUACCACUCC-ACCCGCCCAUUUUUCCUACGGCUCUCUCACA----- ..((((...--(((((((((..(((((((((.....))))))((((....)))).................)))..))-.)))))))..........)))).........----- ( -38.12, z-score = -1.82, R) >droSim1.chrX 2122001 98 + 17042790 ------------GGCGGGGGGCGGGGGUCAUUUAUUAUGGCCAGCUUCUUGGCUUUAACGAAAACCCACUACCACUCCCACCCGCCCAUUUUUCCUUCGGCUCUCUCACA----- ------------(((((((((.(((((((((.....))))))((((....)))).................)).).))).))))))........................----- ( -32.70, z-score = -1.64, R) >droSec1.super_10 2727568 109 + 3036183 UUGCCGGUGGCGGGCGGGGGGCGGGGGUCAUUUAUUAUGGCCAGCUUCUUGGCUUUAAAGAAAACCCACUACCACUCC-ACCCGCCCAUUUUUCCUUCUGCUCUCUCACA----- ......(.((((((.((((((..(((((..(((.(((.((((((....)))))).))).))).)))).)..)).))))-.))))))).......................----- ( -40.40, z-score = -1.61, R) >droEre2.scaffold_4690 421123 100 + 18748788 --------UUGUGGAGGAGGGGGUAGGUCAUUUAUUAUGGCCAGCUUCUUGGCUUUUCCGAAAAACC-----CACUCC-ACUCGCCCAUUUUUCCUUUGACUUGCUCUCUCUCU- --------....((((.(((((((((((((........((((((....)))))).....((((((..-----......-.........))))))...)))))))))))))))))- ( -28.83, z-score = -1.04, R) >droAna3.scaffold_12613 76266 104 - 519072 ----------CUGGAGUUGGGGGCCAGUCAUUUAUUAUGGCCAGCUUCUUGGCUUUUACCUCCCCCC-CUCCCCUACUCACUCCUCUAUCCACCCAACAAAGCCACUGCCUCGCU ----------.(((.(((((((((((...........)))))((((....)))).............-........................))))))....))).......... ( -23.30, z-score = 0.01, R) >consensus ___________GGGAGGGGGGCGGGGGUCAUUUAUUAUGGCCAGCUUCUUGGCUUUAACGAAAACCCACUACCACUCC_ACCCGCCCAUUUUUCCUUCGGCUCUCUCACA_____ ............((....(((((((((((((.....))))))((((....))))..........................)))))))......)).................... (-19.52 = -20.28 + 0.76)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:10:42 2011