| Sequence ID | dm3.chrX |
|---|---|
| Location | 2,959,829 – 2,959,930 |
| Length | 101 |
| Max. P | 0.981756 |

| Location | 2,959,829 – 2,959,930 |
|---|---|
| Length | 101 |
| Sequences | 12 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 93.69 |
| Shannon entropy | 0.14524 |
| G+C content | 0.33826 |
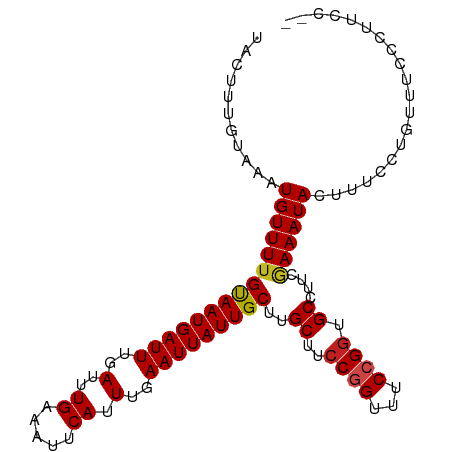
| Mean single sequence MFE | -18.68 |
| Consensus MFE | -16.85 |
| Energy contribution | -16.78 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.981756 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
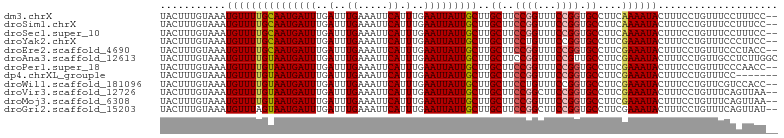
>dm3.chrX 2959829 101 - 22422827 UACUUUGUAAAUGUUUUGCAAUGAUUUGAUUUGAAAUUCAUUUGAAUUAUUGCUUGCUUCCGGUUUCCGGUGCCUUCAAAAUACUUUCCUGUUUCCUUUCC-- ...........(((((((((((((((..(..((.....)).)..)))))))))..((..((((...)))).))....))))))..................-- ( -19.40, z-score = -2.86, R) >droSim1.chrX 2096985 101 - 17042790 UACUUUGUAAAUGUUUUGCAAUGAUUUGAUUUGAAAUUCAUUUGAAUUAUUGCUUGCUUCCGGUUUCCGGUGCCUUCAAAAUACUUUCCUGUUUCCUUUCC-- ...........(((((((((((((((..(..((.....)).)..)))))))))..((..((((...)))).))....))))))..................-- ( -19.40, z-score = -2.86, R) >droSec1.super_10 2698802 101 - 3036183 UACUUUGUAAAUGUUUUGCAAUGAUUUGAUUUGAAAUUCAUUUGAAUUAUUGCUUGCUUCCGGUUUCCGGUGCCUUCAAAAUACUUUCCUGUUUCCUUUCC-- ...........(((((((((((((((..(..((.....)).)..)))))))))..((..((((...)))).))....))))))..................-- ( -19.40, z-score = -2.86, R) >droYak2.chrX 16367135 101 + 21770863 UACUUUGUAAAUGUUUUGCAAUGAUUUGAUUUGAAAUUCAUUUGAAUUAUUGCUUGCUUCCUGUUUCCGGUGCCUUCGAAAUACUUUCCUGUUUCCCUUCC-- .................(((((((((..(..((.....)).)..)))))))))..((..((.......)).))....((((((......))))))......-- ( -16.10, z-score = -2.50, R) >droEre2.scaffold_4690 388083 101 - 18748788 UACUUUGUAAAUGUUUUGCAAUGAUUUGAUUUGAAAUUCAUUUGAAUUAUUGCUUGCUUCCGGUUUCCGGUGCCUUCGAAAUACUUUCCUGUUUCCCUACC-- .................(((((((((..(..((.....)).)..)))))))))..((..((((...)))).))....((((((......))))))......-- ( -20.60, z-score = -3.26, R) >droAna3.scaffold_12613 38377 103 + 519072 UACUUUGUAAAUGUUUUGUAAUGAUUUGAUUUGAAAUUCAUUUGAAUUAUUGCUUGCUUCCGGUUUCCGUUGCCUUCGAAAUACUUUCCUGUUGCCUCUUGGC ...........(((((((((((((((..(..((.....)).)..)))))))))..((...(((...)))..))....))))))..........(((....))) ( -17.20, z-score = -1.64, R) >droPer1.super_18 1317011 101 - 1952607 UACUUUGUAAAUGUUUUGUAAUGAUUUGAUUUGAAAUUCAUUUGAAUUAUUGCUUGCUUCCGGUUUCCGGUGCCUUCGAAAUACUUUCCUGUUUCCCAACC-- .................(((((((((..(..((.....)).)..)))))))))..((..((((...)))).))....((((((......))))))......-- ( -18.60, z-score = -2.37, R) >dp4.chrXL_group1e 2445357 96 - 12523060 UACUUUGUAAAUGUUUUGUAAUGAUUUGAUUUGAAAUUCAUUUGAAUUAUUGCUUGCUUCCGGUUUCCGGUGCCUUCGAAAUACUUUCCUGUUUCC------- .................(((((((((..(..((.....)).)..)))))))))..((..((((...)))).))....((((((......)))))).------- ( -18.60, z-score = -2.82, R) >droWil1.scaffold_181096 10022952 101 + 12416693 UACUUUGUAAAUGUUUUGUAAUGAUUUGAUUUGAAAUUCAUUUGAAUUAUUGCUUGCUUCCUGUUUCCGGUGCCUUCGAAAUACUUUCCUGUUCGUCCACC-- ............((...(((((((((..(..((.....)).)..)))))))))..))...........((((....((((...........))))..))))-- ( -15.70, z-score = -1.82, R) >droVir3.scaffold_12726 327467 101 - 2840439 UACUUUGUAAAUGUUUUGUAAUGAUUUGAUUUGAAAUUCAUUUGAAUUAUUGCUUGCUUCCGGCUUCCGGUGCCUUCGAAAUACUUUCCUGUUUCAGUUAA-- .................(((((((((..(..((.....)).)..)))))))))..((..((((...)))).))....((((((......))))))......-- ( -19.50, z-score = -2.23, R) >droMoj3.scaffold_6308 1996884 101 - 3356042 UACUUUGUAAAUGUUUUGUAAUGAUUUGAUUUGAAAUUCAUUUGAAUUAUUGCUUGCUUCCGGUUUCCGGUGCCUUCGAAAUACUUUCCUGUUUCAGUUAA-- .................(((((((((..(..((.....)).)..)))))))))..((..((((...)))).))....((((((......))))))......-- ( -19.50, z-score = -2.32, R) >droGri2.scaffold_15203 2541886 101 - 11997470 UACUUUGUAAAUGUUUAGUAAUGAUUUGAUUUGAAAUUCAUUUGAAUUAUUGCUUGCUUCCGGCUUCCGGUGCCUUCGAAAUACUUUCCUGUUUCAGUUAU-- ................((((((((((..(..((.....)).)..)))))))))).((..((((...)))).))....((((((......))))))......-- ( -20.20, z-score = -2.23, R) >consensus UACUUUGUAAAUGUUUUGUAAUGAUUUGAUUUGAAAUUCAUUUGAAUUAUUGCUUGCUUCCGGUUUCCGGUGCCUUCGAAAUACUUUCCUGUUUCCCUUCC__ ...........(((((((((((((((..(..((.....)).)..)))))))))..((..((((...)))).))....)))))).................... (-16.85 = -16.78 + -0.08)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:10:40 2011