| Sequence ID | dm3.chrX |
|---|---|
| Location | 2,947,240 – 2,947,340 |
| Length | 100 |
| Max. P | 0.993653 |

| Location | 2,947,240 – 2,947,340 |
|---|---|
| Length | 100 |
| Sequences | 9 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 89.02 |
| Shannon entropy | 0.21933 |
| G+C content | 0.41208 |
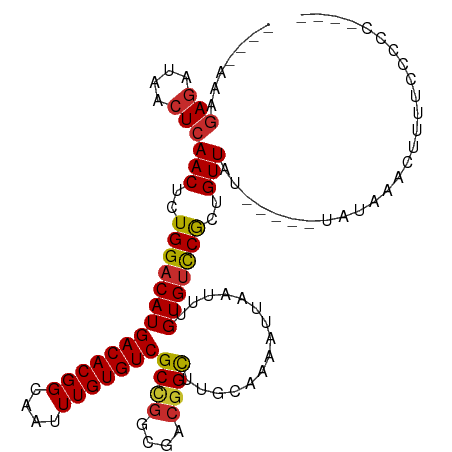
| Mean single sequence MFE | -27.91 |
| Consensus MFE | -25.64 |
| Energy contribution | -25.68 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.63 |
| SVM RNA-class probability | 0.993653 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
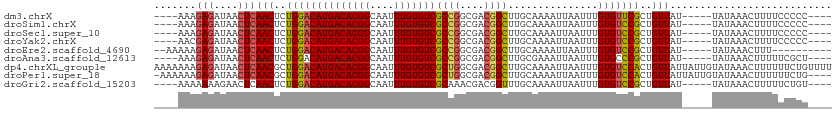
>dm3.chrX 2947240 100 - 22422827 ----AAAGAGAUAACUCAACUCUGGACAUGACACGGCAAUUUGUGUCGCCGGCGACGGCUUGCAAAAUUAAUUUGUGUUCGCUGUUAU-----UAUAAACUUUUCCCCC---- ----...(((....)))(((..((((((((((((((....)))))))((((....))))...............)))))))..)))..-----................---- ( -26.30, z-score = -2.11, R) >droSim1.chrX 2084638 100 - 17042790 ----AAAGAGAUAACUCAACUCUGGACAUGACACGGCAAUUUGUGUCGCCGGCGACGGCUUGCAAAAUUAAUUUGUGUCCGCUGUUAU-----UAUAAACUUUUCCCCC---- ----...(((....)))(((..((((((((((((((....)))))))((((....))))...............)))))))..)))..-----................---- ( -29.20, z-score = -3.07, R) >droSec1.super_10 2686729 100 - 3036183 ----AAAGAGAUAACUCAACUCUGGACAUGACACGGCAAUUUGUGUCGCCGGCGACGGCUUGCAAAAUUAAUUUGUGUCCGCUGUUAU-----UAUAAACUUUUCCCCC---- ----...(((....)))(((..((((((((((((((....)))))))((((....))))...............)))))))..)))..-----................---- ( -29.20, z-score = -3.07, R) >droYak2.chrX 16354699 100 + 21770863 ----AACGAGAUAACUCAACUCUGGACAUGACACGGCAAUUUGUGUCGCCGGCGACGGCUUGCAAAAUUAAUUUGUGUCCGCUGUUAU-----UAUAAACUUUUCCCCC---- ----...(((....)))(((..((((((((((((((....)))))))((((....))))...............)))))))..)))..-----................---- ( -29.30, z-score = -3.08, R) >droEre2.scaffold_4690 375291 96 - 18748788 --AAAAAGAGAUAACUCAACUCUGGACAUGACACGGCAAUUUGUGUCGCCGGCGACGGCUUGCAAAAUUAAUUUGUGUCCGCUGUUAU-----UAUAAACUUU---------- --.....(((....)))(((..((((((((((((((....)))))))((((....))))...............)))))))..)))..-----..........---------- ( -29.20, z-score = -3.47, R) >droAna3.scaffold_12613 21430 100 + 519072 ----AAAGAGAUAACUCAACUCUGGACAUGACACGGCAAUUUGUGUCGCCGGCGACGGCUUGCGAAAUUAAUUUGUGCCCGCUGUUAU-----UAUAAACUUUUUCGCU---- ----...(((....)))............(((((((....)))))))((((....))))..((((((....((((((...........-----))))))...)))))).---- ( -24.50, z-score = -0.90, R) >dp4.chrXL_group1e 2427673 113 - 12523060 AAAAAAAGAGAUAACUCAACGCUGGACAUGACACGGCAAUUUGUGUCGCUGGCGACGGCUUGCAAAAUUAAUUUGUGUCCACUGUUAUUAUUGUAUAAACUUUUUUCUGUUUU .(((((((.((....))((((.((((((((((((((....)))))))((((....))))...............))))))).)))).............)))))))....... ( -28.90, z-score = -2.29, R) >droPer1.super_18 1298511 108 - 1952607 -AAAAAAGAGAUAACUCAACGCUGGACAUGACACGGCAAUUUGUGUCGCUGGCGACGGCUUGCAAAAUUAAUUUGUGUCCACUGUUAUUAUUGUAUAAACUUUUUUCUG---- -(((((((.((....))((((.((((((((((((((....)))))))((((....))))...............))))))).)))).............)))))))...---- ( -28.60, z-score = -2.70, R) >droGri2.scaffold_15203 9526217 100 + 11997470 ----AAAAAAAGAACUCAACUCUGGACAUGACACGGCAAUUUGUGUCGCAAACGACGGUUUGCAAAAUUAAUUUGUGUCCGCUGUUAU-----UAUAAACUUUUUCUGU---- ----..((((((.....(((..((((((((((((((....)))))))((((((....))))))...........)))))))..)))..-----......))))))....---- ( -26.02, z-score = -2.87, R) >consensus ____AAAGAGAUAACUCAACUCUGGACAUGACACGGCAAUUUGUGUCGCCGGCGACGGCUUGCAAAAUUAAUUUGUGUCCGCUGUUAU_____UAUAAACUUUUCCCCC____ .......(((....)))(((..((((((((((((((....)))))))((((....))))...............)))))))..)))........................... (-25.64 = -25.68 + 0.04)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:10:37 2011