
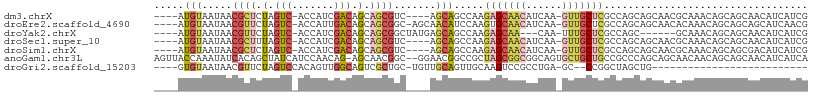
| Sequence ID | dm3.chrX |
|---|---|
| Location | 2,933,405 – 2,933,535 |
| Length | 130 |
| Max. P | 0.856064 |

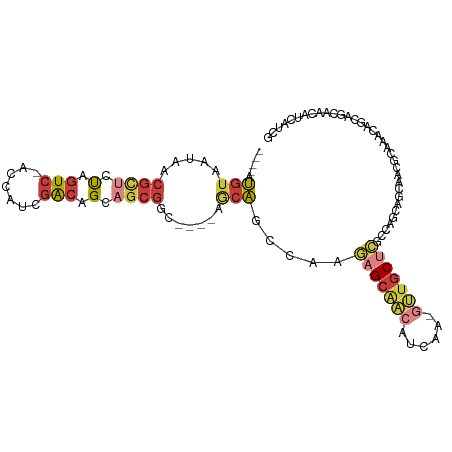
| Location | 2,933,405 – 2,933,504 |
|---|---|
| Length | 99 |
| Sequences | 7 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 66.13 |
| Shannon entropy | 0.66377 |
| G+C content | 0.52310 |
| Mean single sequence MFE | -25.73 |
| Consensus MFE | -7.54 |
| Energy contribution | -8.67 |
| Covariance contribution | 1.13 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.29 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.700737 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 2933405 99 + 22422827 ----AUGUAAUAACGCUCUAGUC-ACCAUCGACAGCAGCGUC----AGCAGCCAAGAGCAACAUCAA-GUUGCUCGCCAGCAGCAACGCAAACAGCAGCAACAUCAUCG ----((((....(((((((.(((-......))))).))))).----.((.((...(((((((.....-)))))))....))......((.....)).)).))))..... ( -27.20, z-score = -3.56, R) >droEre2.scaffold_4690 361181 102 + 18748788 ----AUGUAAUAACGUUCUAGUC-ACCAUUGACAGCAGCGGC-AGCAACAUCCAAGUGCAACAUCAA-GUUGCUCGCCAGCAGCAACACAAACAGCAGCAGCAUCAACG ----.........((((...(((-(....)))).((.((.((-.(((.(......))))........-(((((((....).)))))).......)).)).))...)))) ( -22.10, z-score = -0.82, R) >droYak2.chrX 16340680 94 - 21770863 ----AUGUAAUAACGUUCUAGUC-ACCAUCGACAGCAGCGGCUAUGAGCAGCCAAGAGCAA---CAA-UUUGCUCGCCAGC------GCAAACAGCAGCAACAUCAUCG ----((((............(((-......))).((.((((((......))))..((((((---...-.))))))....))------((.....)).)).))))..... ( -23.10, z-score = -1.27, R) >droSec1.super_10 2673260 99 + 3036183 ----AUGUAAUAACGCUUUAGUC-ACCAUCGACAGCAGCGUC----AGCAGCCAAGAGCAACAUCAA-GUUGCUCGCCAGCAGCAACGCAAACAGCAGCAACAUCAUCG ----.(((......(((...(((-......)))))).((((.----.((.((...(((((((.....-)))))))....)).)).)))).....)))............ ( -25.70, z-score = -3.16, R) >droSim1.chrX 2072200 99 + 17042790 ----AUGUAAUAACGCUCUAGUC-ACCAUCGACAGCAGCGUC----AGCAGCCAAGAGCAACAUCAA-GUUGCUCGCCAGCAGCAACGCAAACAGCAGCGACAUCAUCG ----.........((((((.(((-......)))....((((.----.((.((...(((((((.....-)))))))....)).)).))))....)).))))......... ( -28.80, z-score = -3.44, R) >anoGam1.chr3L 7162761 106 + 41284009 AGUUACCAAAUAUCACAGCUAUCAUCCAACAG-AGCAACGGC--GGAACGGCCGCUAGCGGCGGCAGUGCUGCUGCCGCCCAGCAGCAACAACAGCAGCAACAUCAUCA .................(((.((........)-)((...(((--......)))(((.((((((((((.....))))))))..))))).......))))).......... ( -32.70, z-score = -1.30, R) >droGri2.scaffold_15203 2488070 75 + 11997470 ----GUGUAAUAACGUUCUAGUCCACAGUUGGCAGUCGCUGC-UGUUGCAGUUGCAAGUCCGCCUGA-GC--CCGGCUAGCUG-------------------------- ----.....................((((((((.((.(((((-....))))).))......((....-))--...))))))))-------------------------- ( -20.50, z-score = 0.38, R) >consensus ____AUGUAAUAACGCUCUAGUC_ACCAUCGACAGCAGCGGC____AGCAGCCAAGAGCAACAUCAA_GUUGCUCGCCAGCAGCAACGCAAACAGCAGCAACAUCAUCG .............((((.(.(((.......))).).))))...............(((((((......))))))).................................. ( -7.54 = -8.67 + 1.13)

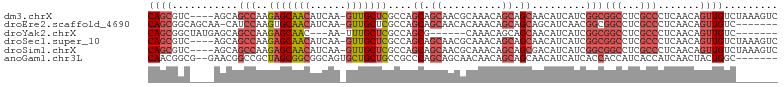
| Location | 2,933,435 – 2,933,535 |
|---|---|
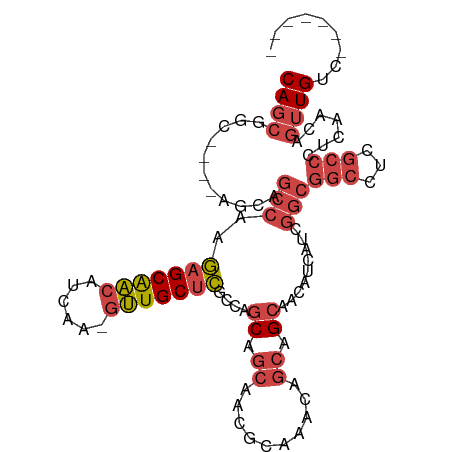
| Length | 100 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 76.41 |
| Shannon entropy | 0.44020 |
| G+C content | 0.57791 |
| Mean single sequence MFE | -28.57 |
| Consensus MFE | -14.76 |
| Energy contribution | -16.23 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.856064 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 2933435 100 + 22422827 CAGCGUC----AGCAGCCAAGAGCAACAUCAA-GUUGCUCGCCAGCAGCAACGCAAACAGCAGCAACAUCAUCGGCGGCCUCGCCCUCAACAGUUGUCUAAAGUC ..((((.----.((.((...(((((((.....-)))))))....)).)).))))........(((((......((((....)))).......)))))........ ( -29.12, z-score = -3.02, R) >droEre2.scaffold_4690 361211 96 + 18748788 CAGCGGCAGCAA-CAUCCAAGUGCAACAUCAA-GUUGCUCGCCAGCAGCAACACAAACAGCAGCAGCAUCAACGGCGGCCUCGCCCUCAACAGUUGUC------- ..((((((((((-(..................-)))))).))).)).(((((.......((....))......((((....)))).......))))).------- ( -25.87, z-score = -1.30, R) >droYak2.chrX 16340710 88 - 21770863 CAGCGGCUAUGAGCAGCCAAGAGCAAC---AA-UUUGCUCGCCAGCG------CAAACAGCAGCAACAUCAUCGGCGGCCUCGCCCUCAACAGUUGUC------- ..((((((.((((..(((..((((((.---..-.))))))....(((------(.....)).)).........)))(((...)))))))..)))))).------- ( -26.60, z-score = -0.97, R) >droSec1.super_10 2673290 100 + 3036183 CAGCGUC----AGCAGCCAAGAGCAACAUCAA-GUUGCUCGCCAGCAGCAACGCAAACAGCAGCAACAUCAUCGGCGGCCUCGCCCUCAACAGUUGUCUAAAGUC ..((((.----.((.((...(((((((.....-)))))))....)).)).))))........(((((......((((....)))).......)))))........ ( -29.12, z-score = -3.02, R) >droSim1.chrX 2072230 100 + 17042790 CAGCGUC----AGCAGCCAAGAGCAACAUCAA-GUUGCUCGCCAGCAGCAACGCAAACAGCAGCGACAUCAUCGGCGGCCUCGCCCUCAACAGUUGUCUAAAGUC ..((((.----.((.((...(((((((.....-)))))))....)).)).)))).....(((((((.....))((((....)))).......)))))........ ( -29.70, z-score = -2.50, R) >anoGam1.chr3L 7162795 96 + 41284009 CAACGGCG--GAACGGCCGCUAGCGGCGGCAGUGCUGCUGCCGCCCAGCAGCAACAACAGCAGCAACAUCAUCACCACCAUCACCAUCAACUACUGGC------- ....(((.--.....)))(((((.((((((((.....))))))))..((.((.......)).)).............................)))))------- ( -31.00, z-score = -0.81, R) >consensus CAGCGGC____AGCAGCCAAGAGCAACAUCAA_GUUGCUCGCCAGCAGCAACGCAAACAGCAGCAACAUCAUCGGCGGCCUCGCCCUCAACAGUUGUC_______ ((((...........(((..(((((((......)))))))....((.((..........)).)).........)))(((...))).......))))......... (-14.76 = -16.23 + 1.48)
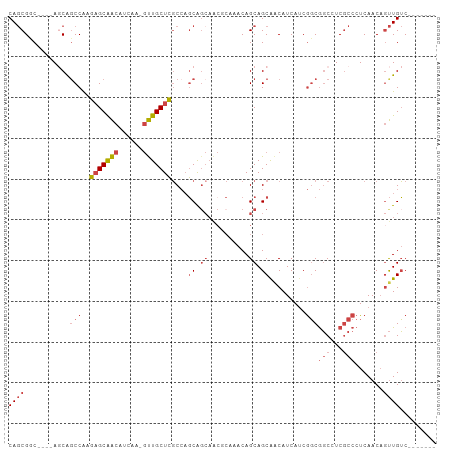
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:10:33 2011