| Sequence ID | dm3.chrX |
|---|---|
| Location | 2,932,266 – 2,932,400 |
| Length | 134 |
| Max. P | 0.982788 |

| Location | 2,932,266 – 2,932,400 |
|---|---|
| Length | 134 |
| Sequences | 6 |
| Columns | 135 |
| Reading direction | forward |
| Mean pairwise identity | 85.21 |
| Shannon entropy | 0.28594 |
| G+C content | 0.42006 |
| Mean single sequence MFE | -34.53 |
| Consensus MFE | -21.66 |
| Energy contribution | -24.80 |
| Covariance contribution | 3.14 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
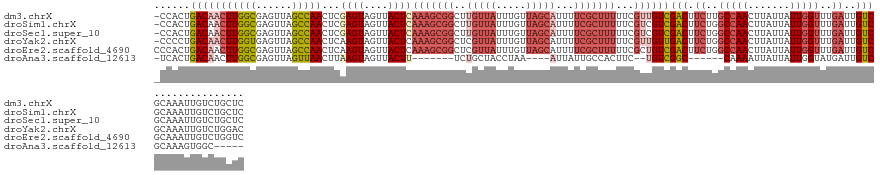
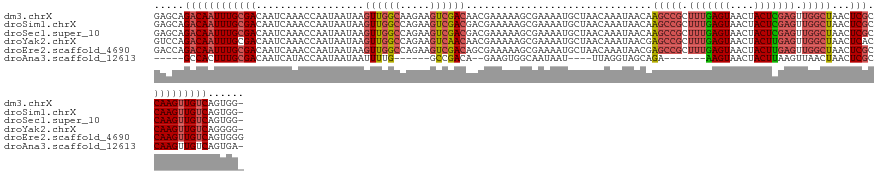
>dm3.chrX 2932266 134 + 22422827 -CCACUGACAACUUGGCGAGUUAGCCAACUCGAGUAGUUACUCAAAGCGGCUUGUUAUUUGUUAGCAUUUUCGCUUUUUCGUUGUCGACUUCUUGCCAACUUAUUAUUGGUUUGAUUGUCGCAAAUUGUCUGCUC -.....(((((((((((......)))))...((((....))))(((((((..(((((.....)))))...)))))))...))))))(((.((..(((((.......)))))..))..)))(((.......))).. ( -39.60, z-score = -3.23, R) >droSim1.chrX 2071061 134 + 17042790 -CCACUGACAACUUGGCGAGUUAGCCAACUCGAGUAGUUACUCAAAGCGGCUUGUUAUUUGUUAGCAUUUUCGCUUUUUCGUCGUCGACUUCUGGCCAACUUAUUAUUGGUUUGAUUGUCGCAAAUUGUCUGCUC -.....(((((...((((((((....)))))((((....))))(((((((..(((((.....)))))...)))))))...)))(.((((.((..(((((.......)))))..))..)))))...)))))..... ( -37.00, z-score = -2.07, R) >droSec1.super_10 2672122 134 + 3036183 -CCACUGACAACUUGGCGAGUUAGCCAACUCGAGUAGUUACUCAAAGCGGCUUGUUAUUUGUUAGCAUUUUCGCUUUUUCGUCGUCGACUUCUGGCCAACUUAUUAUUGGUUUGAUUGUCGCAAAUUGUCUGCUC -.....(((((...((((((((....)))))((((....))))(((((((..(((((.....)))))...)))))))...)))(.((((.((..(((((.......)))))..))..)))))...)))))..... ( -37.00, z-score = -2.07, R) >droYak2.chrX 16339557 134 - 21770863 -CCCCUGACAACUUGGUGAGUUAGCCAACUCAAGUAGUUACUCAAAGCGGCUCGUUAUUUGUUAGCAUUUUCGCUUUUUCGUUGUUGACUUCUGGCCAACUUAUUAUUGGUUUGAUUGUCGCAAAUUGUCUGGAC -..((.(((((((...((((((....))))))...)))).......(((((.........(((((((....((......)).))))))).((..(((((.......)))))..))..))))).....))).)).. ( -34.10, z-score = -1.33, R) >droEre2.scaffold_4690 360048 135 + 18748788 CCCACUGACAACUUGGCGAGUUAGCCAACUCAAGUAGUUACUCAAAGCGGCUCGUUAUUUGUUAGCAUUUUCGCUUUUUCGCUGUCGACUUCUGGCCAACUUAUUAUUGGUUUGAUUGUCGCAAAUUGUCUGGUC .(((..(((((.((((((((..(((........(.(((..(.....)..))))((((.....))))......)))..)))))...((((.((..(((((.......)))))..))..))))))).)))))))).. ( -34.70, z-score = -1.32, R) >droAna3.scaffold_12613 1675 110 - 519072 -UCACUGACAACUUGGCGAGUUAGUUAACUUAAGUAGUUACUU-------UCUGCUACCUAA----AUUAUUGCCACUUC--UGUCGGC------CAAAAUUAUUAUUGGUAUGAUUGUCGCAAAGUGGC----- -............((((.((..(((.((((.....))))))).-------.)))))).....----......(((((((.--((.((((------...((((((.......)))))))))))))))))))----- ( -24.80, z-score = -0.93, R) >consensus _CCACUGACAACUUGGCGAGUUAGCCAACUCAAGUAGUUACUCAAAGCGGCUUGUUAUUUGUUAGCAUUUUCGCUUUUUCGUUGUCGACUUCUGGCCAACUUAUUAUUGGUUUGAUUGUCGCAAAUUGUCUGCUC ......(((((((((((......)))))...((((....))))(((((((..(((((.....)))))...)))))))...))))))(((.((..(((((.......)))))..))..)))............... (-21.66 = -24.80 + 3.14)
| Location | 2,932,266 – 2,932,400 |
|---|---|
| Length | 134 |
| Sequences | 6 |
| Columns | 135 |
| Reading direction | reverse |
| Mean pairwise identity | 85.21 |
| Shannon entropy | 0.28594 |
| G+C content | 0.42006 |
| Mean single sequence MFE | -34.99 |
| Consensus MFE | -25.89 |
| Energy contribution | -27.25 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.11 |
| SVM RNA-class probability | 0.982788 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
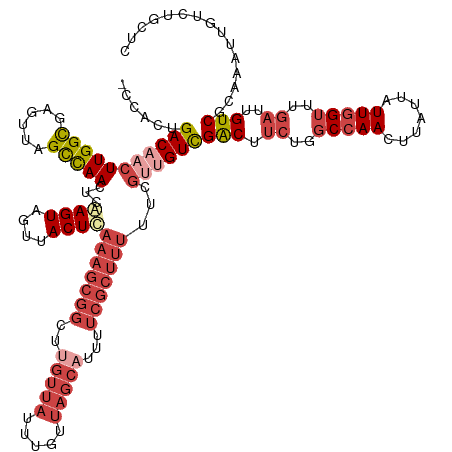
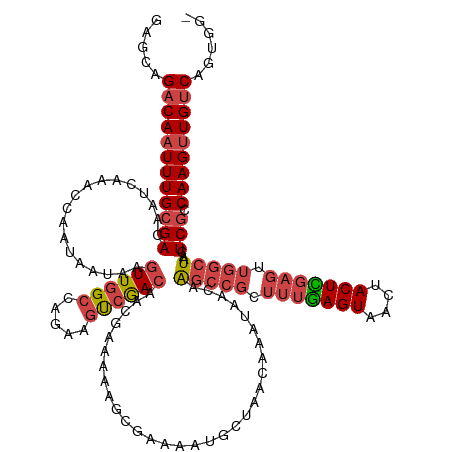
>dm3.chrX 2932266 134 - 22422827 GAGCAGACAAUUUGCGACAAUCAAACCAAUAAUAAGUUGGCAAGAAGUCGACAACGAAAAAGCGAAAAUGCUAACAAAUAACAAGCCGCUUUGAGUAACUACUCGAGUUGGCUAACUCGCCAAGUUGUCAGUGG- .....((((((((((((..................(((((((.....(((.(.........))))...)))))))........(((((.(((((((....))))))).)))))...))).))))))))).....- ( -37.80, z-score = -3.27, R) >droSim1.chrX 2071061 134 - 17042790 GAGCAGACAAUUUGCGACAAUCAAACCAAUAAUAAGUUGGCCAGAAGUCGACGACGAAAAAGCGAAAAUGCUAACAAAUAACAAGCCGCUUUGAGUAACUACUCGAGUUGGCUAACUCGCCAAGUUGUCAGUGG- .....((((((((((((..................((((((......(((.(.........))))....))))))........(((((.(((((((....))))))).)))))...))).))))))))).....- ( -36.90, z-score = -2.73, R) >droSec1.super_10 2672122 134 - 3036183 GAGCAGACAAUUUGCGACAAUCAAACCAAUAAUAAGUUGGCCAGAAGUCGACGACGAAAAAGCGAAAAUGCUAACAAAUAACAAGCCGCUUUGAGUAACUACUCGAGUUGGCUAACUCGCCAAGUUGUCAGUGG- .....((((((((((((..................((((((......(((.(.........))))....))))))........(((((.(((((((....))))))).)))))...))).))))))))).....- ( -36.90, z-score = -2.73, R) >droYak2.chrX 16339557 134 + 21770863 GUCCAGACAAUUUGCGACAAUCAAACCAAUAAUAAGUUGGCCAGAAGUCAACAACGAAAAAGCGAAAAUGCUAACAAAUAACGAGCCGCUUUGAGUAACUACUUGAGUUGGCUAACUCACCAAGUUGUCAGGGG- .(((.(((((((((((...................((((((.....))))))........(((......))).........))(((((.((..(((....)))..)).))))).......))))))))).))).- ( -33.60, z-score = -2.12, R) >droEre2.scaffold_4690 360048 135 - 18748788 GACCAGACAAUUUGCGACAAUCAAACCAAUAAUAAGUUGGCCAGAAGUCGACAGCGAAAAAGCGAAAAUGCUAACAAAUAACGAGCCGCUUUGAGUAACUACUUGAGUUGGCUAACUCGCCAAGUUGUCAGUGGG ..((((((((((((((((..((...(((((.....)))))...)).))))...((((...(((......)))...........(((((.((..(((....)))..)).)))))...)))))))))))))..))). ( -39.20, z-score = -3.12, R) >droAna3.scaffold_12613 1675 110 + 519072 -----GCCACUUUGCGACAAUCAUACCAAUAAUAAUUUUG------GCCGACA--GAAGUGGCAAUAAU----UUAGGUAGCAGA-------AAGUAACUACUUAAGUUAACUAACUCGCCAAGUUGUCAGUGA- -----((((((((.((.(((.................)))------..))...--))))))))..((((----((((((((....-------......))))))))))))..(((((.....))))).......- ( -25.53, z-score = -1.71, R) >consensus GAGCAGACAAUUUGCGACAAUCAAACCAAUAAUAAGUUGGCCAGAAGUCGACAACGAAAAAGCGAAAAUGCUAACAAAUAACAAGCCGCUUUGAGUAACUACUCGAGUUGGCUAACUCGCCAAGUUGUCAGUGG_ .....((((((((((((..................((((((.....))))))...............................(((((.(((((((....))))))).)))))...))).)))))))))...... (-25.89 = -27.25 + 1.36)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:10:31 2011