
| Sequence ID | dm3.chrX |
|---|---|
| Location | 2,887,779 – 2,887,879 |
| Length | 100 |
| Max. P | 0.948224 |

| Location | 2,887,779 – 2,887,879 |
|---|---|
| Length | 100 |
| Sequences | 11 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 80.50 |
| Shannon entropy | 0.39856 |
| G+C content | 0.40958 |
| Mean single sequence MFE | -26.26 |
| Consensus MFE | -15.65 |
| Energy contribution | -16.41 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.857396 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
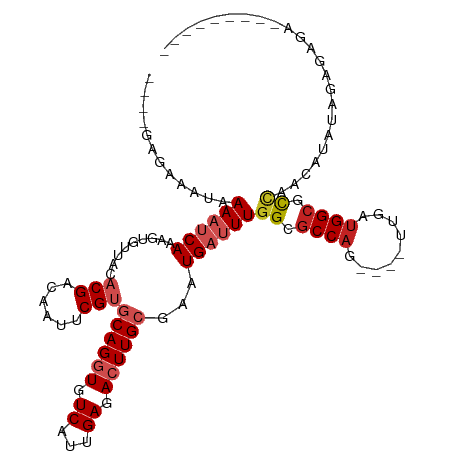
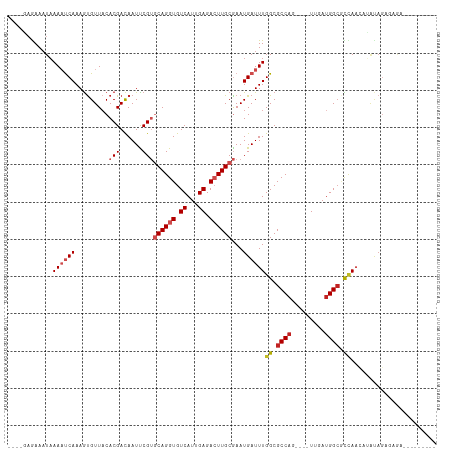
>dm3.chrX 2887779 100 + 22422827 ----GAGAAAUAAAAUCAAAGUGUUACACGACAAUCCGUGCAGGUGUCAUUGAGACUUGCGAAUGAUUUGGCGCCAG----UUGAUGGCGCCAACAUAUAGACAGAUU------- ----........((((((.........(((......)))((((((.((...)).))))))...))))))(((((((.----....)))))))................------- ( -29.40, z-score = -2.72, R) >droSim1.chrU 12452289 100 + 15797150 ----GCGAAAUAAAAUCAAAGUGUUACACGACAAUCCGUGCAGGUGUCAUUGAGACUUGCGAAUGAUUUGGCGCCAG----UUGAUGGCGCCAACAUAUAGAGAGAUU------- ----........((((((.........(((......)))((((((.((...)).))))))...))))))(((((((.----....)))))))................------- ( -29.40, z-score = -2.37, R) >droSec1.super_10 2628540 100 + 3036183 ----GCAAAAUAAAAUCAAAGUGUUACACGACAAUCCGUGCAGGUGUCAUUGAGACUUGCGAAUGAUUUGGCGCCAA----UUGAUGGCGCCAACAUAUAGAGAGAUU------- ----........((((((.........(((......)))((((((.((...)).))))))...))))))(((((((.----....)))))))................------- ( -28.60, z-score = -2.47, R) >droYak2.chrX 16295564 106 - 21770863 ----GAGAAAUAAAAUCAAAGUGUUACACGACAAUCCGUGCAGGUGUCAUUGAGACUUGCGAAUGAUUUGGCGCCAGCCAGUUGAUGGCGCCAACAUAUAGCGAGAGAAU----- ----...........((((.(((...)))((((..((.....)))))).))))..(((((..(((..(((((((((..(....).))))))))))))...))))).....----- ( -31.30, z-score = -2.63, R) >droEre2.scaffold_4690 313655 107 + 18748788 ----GAGAAAUAAAAUCAAAGUGUUACACGACAAUCCGUGCAGGUGUCAUUGAGACUUGCGAAUGAUUUGGCGCCAG----UUGAUGGCGCCAACAUAUAGAGAAUAGAGAGAAU ----........((((((.........(((......)))((((((.((...)).))))))...))))))(((((((.----....)))))))....................... ( -29.40, z-score = -3.07, R) >droAna3.scaffold_13248 4779569 94 - 4840945 AAAUAAGAAAUAAAAUCAAAGUGUUACACGACAAUUCGUGCAGGUGUCAUUGAGACUUGCGAAUGAUUUGGCGCCAG----UUAAUGGCGCUAAGUUU----------------- ...............((..........((((....))))((((((.((...)).))))))))..((((((((((((.----....)))))))))))).----------------- ( -28.90, z-score = -3.36, R) >dp4.chrXL_group1e 2353014 96 + 12523060 ----AAGAAAUAAAACCAAAGUGUUACACGACAAUUCG--CAGGUGUCAUUGAGAGUUGCAAAUGAUUUGGCGCCAG----UUUAUGGCCUCAACUGACAGUUGGA--------- ----...........((((..(((((...(.((((((.--(((......))).))))))).......((((.((((.----....)))).)))).))))).)))).--------- ( -22.90, z-score = -0.50, R) >droPer1.super_18 1223414 96 + 1952607 ----AAGAAAUAAAACCAAAGUGUUACACGACAAUUCG--CAGGUGUCAUUGAGAGUUGCAAAUGAUUUGGCGCCAG----UUUAUGGCCUCAACUGACAGUUGGA--------- ----...........((((..(((((...(.((((((.--(((......))).))))))).......((((.((((.----....)))).)))).))))).)))).--------- ( -22.90, z-score = -0.50, R) >droWil1.scaffold_181096 10117180 98 - 12416693 ----CAAAAUUCAAAUCAAAAUGUUUCACGACAAUUCGUGCAGGUGUCAUUGAGACUUGCGAAUGAUUUGGCGCCAC----UUUAUGGCAUUAAAUGGCAGCUAAA--------- ----.......(((((((.........((((....))))((((((.((...)).))))))...)))))))((((((.----((((......)))))))).))....--------- ( -26.30, z-score = -1.87, R) >droMoj3.scaffold_6308 1873766 101 + 3356042 ----UAAAAAUAAAUUCAAAGCGUUACACGACAAUUCGUGCAGGUGUCAUUGAGACUUGCGAAUGAUUUGGCGCCAG----UUUAUGGCAUUAUACUAUAUGACAGACU------ ----...........(((((.((((.(((((....))))((((((.((...)).))))))))))).))))).((((.----....))))....................------ ( -21.80, z-score = -0.73, R) >droGri2.scaffold_15203 2441896 94 + 11997470 ----CAAAAAUAAAUUCAAAGUGUUACACGAUAAUUCGUGCAGGUGUCAUUGAGACUUGCGAAUGAUUUGGCGCCAG----UUUACGGAAUAAUAUGAUAGU------------- ----.........((((...(((...(((((....)))))..(((((((...................)))))))..----..))).))))...........------------- ( -17.91, z-score = -0.09, R) >consensus ____GAGAAAUAAAAUCAAAGUGUUACACGACAAUUCGUGCAGGUGUCAUUGAGACUUGCGAAUGAUUUGGCGCCAG____UUGAUGGCGCCAACAUAUAGAGAGA_________ ............((((((.........(((......)))((((((.((...)).))))))...))))))((.((((.........)))).))....................... (-15.65 = -16.41 + 0.76)

| Location | 2,887,779 – 2,887,879 |
|---|---|
| Length | 100 |
| Sequences | 11 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 80.50 |
| Shannon entropy | 0.39856 |
| G+C content | 0.40958 |
| Mean single sequence MFE | -23.65 |
| Consensus MFE | -14.23 |
| Energy contribution | -14.69 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.948224 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
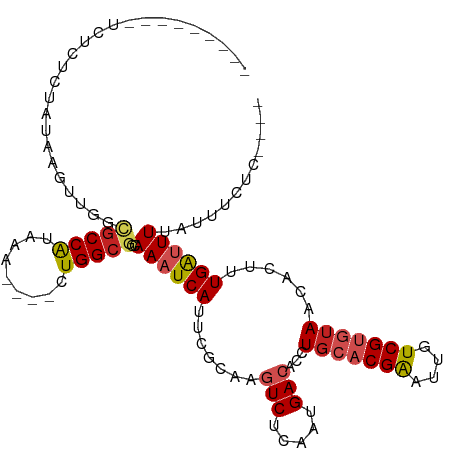
>dm3.chrX 2887779 100 - 22422827 -------AAUCUGUCUAUAUGUUGGCGCCAUCAA----CUGGCGCCAAAUCAUUCGCAAGUCUCAAUGACACCUGCACGGAUUGUCGUGUAACACUUUGAUUUUAUUUCUC---- -------(((((((..((...(((((((((....----.)))))))))...))..))).(((.....)))...(((((((....))))))).......)))).........---- ( -27.40, z-score = -3.01, R) >droSim1.chrU 12452289 100 - 15797150 -------AAUCUCUCUAUAUGUUGGCGCCAUCAA----CUGGCGCCAAAUCAUUCGCAAGUCUCAAUGACACCUGCACGGAUUGUCGUGUAACACUUUGAUUUUAUUUCGC---- -------..............(((((((((....----.)))))))))((((.......(((.....)))...(((((((....)))))))......))))..........---- ( -27.00, z-score = -2.83, R) >droSec1.super_10 2628540 100 - 3036183 -------AAUCUCUCUAUAUGUUGGCGCCAUCAA----UUGGCGCCAAAUCAUUCGCAAGUCUCAAUGACACCUGCACGGAUUGUCGUGUAACACUUUGAUUUUAUUUUGC---- -------..............(((((((((....----.))))))))).......(((((((.....)))...(((((((....)))))))................))))---- ( -29.20, z-score = -3.47, R) >droYak2.chrX 16295564 106 + 21770863 -----AUUCUCUCGCUAUAUGUUGGCGCCAUCAACUGGCUGGCGCCAAAUCAUUCGCAAGUCUCAAUGACACCUGCACGGAUUGUCGUGUAACACUUUGAUUUUAUUUCUC---- -----........((...((((((((((((((....)).)))))))))..)))..))..(((.....)))...(((((((....)))))))....................---- ( -28.90, z-score = -2.67, R) >droEre2.scaffold_4690 313655 107 - 18748788 AUUCUCUCUAUUCUCUAUAUGUUGGCGCCAUCAA----CUGGCGCCAAAUCAUUCGCAAGUCUCAAUGACACCUGCACGGAUUGUCGUGUAACACUUUGAUUUUAUUUCUC---- .....................(((((((((....----.)))))))))((((.......(((.....)))...(((((((....)))))))......))))..........---- ( -27.00, z-score = -3.75, R) >droAna3.scaffold_13248 4779569 94 + 4840945 -----------------AAACUUAGCGCCAUUAA----CUGGCGCCAAAUCAUUCGCAAGUCUCAAUGACACCUGCACGAAUUGUCGUGUAACACUUUGAUUUUAUUUCUUAUUU -----------------.......((((((....----.)))))).((((((.......(((.....)))...(((((((....)))))))......))))))............ ( -23.90, z-score = -3.72, R) >dp4.chrXL_group1e 2353014 96 - 12523060 ---------UCCAACUGUCAGUUGAGGCCAUAAA----CUGGCGCCAAAUCAUUUGCAACUCUCAAUGACACCUG--CGAAUUGUCGUGUAACACUUUGGUUUUAUUUCUU---- ---------.((((.(((((.(((((((((....----.))))((.((....)).))....))))))))))...(--(((....))))........))))...........---- ( -20.60, z-score = -0.88, R) >droPer1.super_18 1223414 96 - 1952607 ---------UCCAACUGUCAGUUGAGGCCAUAAA----CUGGCGCCAAAUCAUUUGCAACUCUCAAUGACACCUG--CGAAUUGUCGUGUAACACUUUGGUUUUAUUUCUU---- ---------.((((.(((((.(((((((((....----.))))((.((....)).))....))))))))))...(--(((....))))........))))...........---- ( -20.60, z-score = -0.88, R) >droWil1.scaffold_181096 10117180 98 + 12416693 ---------UUUAGCUGCCAUUUAAUGCCAUAAA----GUGGCGCCAAAUCAUUCGCAAGUCUCAAUGACACCUGCACGAAUUGUCGUGAAACAUUUUGAUUUGAAUUUUG---- ---------....((.(((((((.........))----)))))))(((((((.......(((.....))).....(((((....)))))........))))))).......---- ( -23.40, z-score = -1.67, R) >droMoj3.scaffold_6308 1873766 101 - 3356042 ------AGUCUGUCAUAUAGUAUAAUGCCAUAAA----CUGGCGCCAAAUCAUUCGCAAGUCUCAAUGACACCUGCACGAAUUGUCGUGUAACGCUUUGAAUUUAUUUUUA---- ------....................((((....----.))))......(((...((..(((.....)))...(((((((....)))))))..))..)))...........---- ( -19.40, z-score = -0.59, R) >droGri2.scaffold_15203 2441896 94 - 11997470 -------------ACUAUCAUAUUAUUCCGUAAA----CUGGCGCCAAAUCAUUCGCAAGUCUCAAUGACACCUGCACGAAUUAUCGUGUAACACUUUGAAUUUAUUUUUG---- -------------...............(((...----...))).......((((....(((.....)))...(((((((....))))))).......)))).........---- ( -12.80, z-score = -0.39, R) >consensus _________UCUCUCUAUAAGUUGGCGCCAUAAA____CUGGCGCCAAAUCAUUCGCAAGUCUCAAUGACACCUGCACGAAUUGUCGUGUAACACUUUGAUUUUAUUUCUC____ .........................(((((.........)))))..((((((.......(((.....)))...(((((((....)))))))......))))))............ (-14.23 = -14.69 + 0.47)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:10:28 2011