| Sequence ID | dm3.chrX |
|---|---|
| Location | 2,870,516 – 2,870,622 |
| Length | 106 |
| Max. P | 0.906894 |

| Location | 2,870,516 – 2,870,622 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 81.32 |
| Shannon entropy | 0.33416 |
| G+C content | 0.43716 |
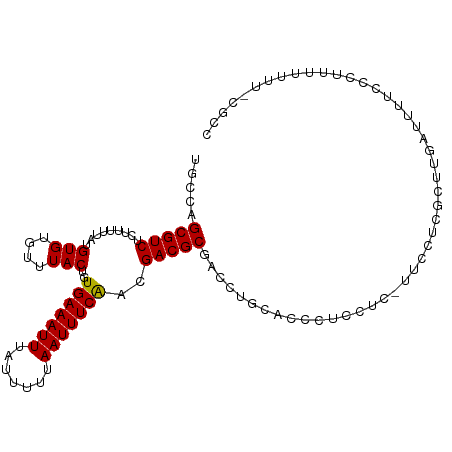
| Mean single sequence MFE | -16.70 |
| Consensus MFE | -12.79 |
| Energy contribution | -12.57 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.906894 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
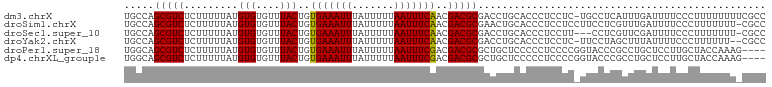
>dm3.chrX 2870516 106 + 22422827 UGCCAGCGUCUCUUUUUAUGUGUGUUUACUGUGAAAUUUAUUUUUAAUUUCAACGACGCGACCUGCACCCUCCUC-UGCCUCAUUUGAUUUUCCCUUUUUUUUCGCC .((..(((((.........(((....)))..(((((((.......)))))))..)))))((...(((........-))).))......................)). ( -14.60, z-score = -1.76, R) >droSim1.chrX 2012921 106 + 17042790 UGCCAGCGUCUCUUUUUAUGUGUGUUUACUGUGAAAUUUAUUUUUAAUUUCAACGACGCGAACUGCACCCUCCUCCUUCCUCGUUUGAUUUUCCCUUUUUUU-CGCC (((..(((((.........(((....)))..(((((((.......)))))))..))))).....)))...................................-.... ( -13.80, z-score = -1.42, R) >droSec1.super_10 2611743 103 + 3036183 UGCCAGCGUCUCUUUUUAUGUGUGUUUACUGUGAAAUUUAUUUUUAAUUUCAACGACGCGACCUGCACCCUCCUU---CCUCGUUCGAUUUUCCCUUUUUUU-CGCC (((..(((((.........(((....)))..(((((((.......)))))))..))))).....)))........---........................-.... ( -13.80, z-score = -1.29, R) >droYak2.chrX 16278475 104 - 21770863 UGCCAGCGUCUCUUUUUAUGUGUGUUUACUGUGAAAUUUAUUUUUAAUUUCAACGACGCGACCUGCACCCUCCUC-UUCCUAGCUUUAUUUUCCCUUUUUU--CGCC (((..(((((.........(((....)))..(((((((.......)))))))..))))).....)))........-.........................--.... ( -13.80, z-score = -1.82, R) >droPer1.super_18 1199850 103 + 1952607 UGGCAGCGUCUCUUUUUAUGUGUGUUUACUGUGAAAUUUAUUUUUAAUUUCGACGACGCGCUGCUCCCCCUCCCCGGUACCCGCCUGCUCCUUGCUACCAAAG---- .((((((((.((.......(((....))).((((((((.......)))))).)))).))))))))..........((((.......((.....))))))....---- ( -22.11, z-score = -2.05, R) >dp4.chrXL_group1e 2327872 103 + 12523060 UGGCAGCGUCUCUUUUUAUGUGUGUUUACUGUGAAAUUUAUUUUUAAUUUCGACGACGCGCUGCUCCCCCUCCCCGGUACCCGCCUGCUCCUUGCUACCAAAG---- .((((((((.((.......(((....))).((((((((.......)))))).)))).))))))))..........((((.......((.....))))))....---- ( -22.11, z-score = -2.05, R) >consensus UGCCAGCGUCUCUUUUUAUGUGUGUUUACUGUGAAAUUUAUUUUUAAUUUCAACGACGCGACCUGCACCCUCCUC_UUCCUCGCUUGAUUUUCCCUUUUUUU_CGCC .....(((((.........(((....)))..(((((((.......)))))))..)))))................................................ (-12.79 = -12.57 + -0.22)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:10:26 2011