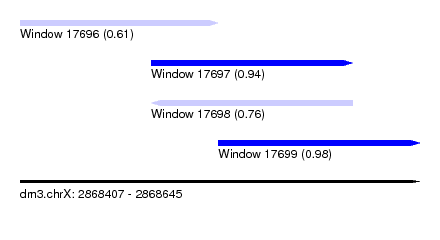
| Sequence ID | dm3.chrX |
|---|---|
| Location | 2,868,407 – 2,868,645 |
| Length | 238 |
| Max. P | 0.980831 |

| Location | 2,868,407 – 2,868,525 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.89 |
| Shannon entropy | 0.11106 |
| G+C content | 0.47120 |
| Mean single sequence MFE | -33.14 |
| Consensus MFE | -30.16 |
| Energy contribution | -30.04 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.91 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.614542 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 2868407 118 + 22422827 CAGAUUGAGAUAAAGUAGGAGAAUUGGCCGUCAAGAGGUCAAUGCUGUCGGGACAGCAGCUGUUCACCA--AUCUCUCCUCCUUCUUCGCACCCACAAUUCUUCCCGUUUUGGUAGAAAU ..(..((.((..(((.(((((((((((((.......)))))))(((((....)))))............--...)))))).)))..)).))..)....((((.((......)).)))).. ( -32.70, z-score = -1.08, R) >droSim1.chrX 2010822 120 + 17042790 CAGAUUGAGAUAAAGUAGGAGAAUUGGCCGUCAAGAGGUCAAUGCUGUCGGGACAGCAGCUGUUCACCAGCAUCUCUCCUCCUUCUUCGCACCCACAAUUCUUCCCGUUUUGGUAGAAAU ..(..((.((..(((.(((((((((((((.......)))))))((((...((((((...))))))..))))...)))))).)))..)).))..)....((((.((......)).)))).. ( -37.20, z-score = -2.08, R) >droSec1.super_10 2609631 120 + 3036183 CAGAUUGAGAUAAAGUAGGAGAAUUGGCCGUCAAGAGGUCAAUGCUGUCGGGACAGCAGCUGUUCACCACCAGCUCUCCUCUUUUUUCGCACCCACAAUUCUUCCCGUUUUGGUAGAAAU ......((((.((((.(((((((((((((.......)))))))(((((....))))).((((........))))))))))))))))))..........((((.((......)).)))).. ( -36.60, z-score = -2.06, R) >droYak2.chrX 16276351 117 - 21770863 CAGAUUAAGAUAAAGUAGGAGAAUUGGCCGUCAAGAGGUCAAUGCUGUCGGGACAGCAGCUGUUCACUAU---CUCUCCUCCUUCCUCGCACCCACAAUUCUUCUCAUUUUGGUAGAAAU ........((..(((.(((((((((((((.......)))))))(((((....))))).............---.)))))).)))..))..........((((.((......)).)))).. ( -29.20, z-score = -0.57, R) >droEre2.scaffold_4690 293755 117 + 18748788 CAGAUUGAGAUAAAGUAGGAGAAUUGGCCGUCAAGAGGUCAAUGCUGUCGGUACAGCAGCUGUUCUCCAU---UUCUCCUCCCUCUUCGCACCCACAAUUCUUCCCGUUUUGGUAGAAAU .(((..(((...((((.((((((((((((.......))))).((((((....))))))...)))))))))---))...)))..)))............((((.((......)).)))).. ( -30.00, z-score = -0.93, R) >consensus CAGAUUGAGAUAAAGUAGGAGAAUUGGCCGUCAAGAGGUCAAUGCUGUCGGGACAGCAGCUGUUCACCAU_A_CUCUCCUCCUUCUUCGCACCCACAAUUCUUCCCGUUUUGGUAGAAAU ........((..(((.(((((((((((((.......)))))))(((((....))))).................)))))).)))..))..........((((.((......)).)))).. (-30.16 = -30.04 + -0.12)

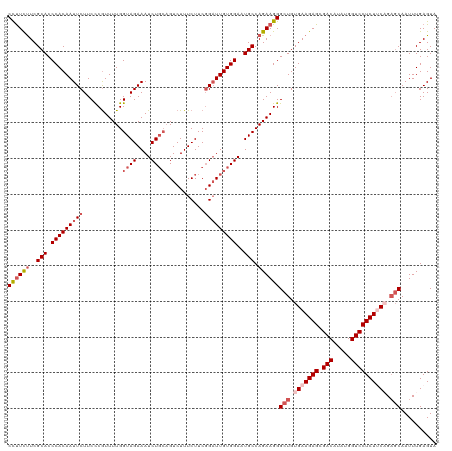
| Location | 2,868,485 – 2,868,605 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.25 |
| Shannon entropy | 0.10374 |
| G+C content | 0.46969 |
| Mean single sequence MFE | -38.47 |
| Consensus MFE | -33.95 |
| Energy contribution | -36.03 |
| Covariance contribution | 2.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.941192 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 2868485 120 + 22422827 CCUUCUUCGCACCCACAAUUCUUCCCGUUUUGGUAGAAAUCUGCAGUCUUUCUUCCGAUUGUGGAAUGCCAGAAGGGCUGUGAAGAGCGACAUAUGGUCCUCUACAGAGCAAUUUGAGUA ((((((..(((.......((((.((......)).)))).((..(((((........)))))..)).))).))))))(((.((.((((.(((.....))))))).)).))).......... ( -39.00, z-score = -2.69, R) >droSim1.chrX 2010902 120 + 17042790 CCUUCUUCGCACCCACAAUUCUUCCCGUUUUGGUAGAAAUCUGCAGUCUUUCUUCGGAUUGUGGAAUGCCAGAAGGGCUGUGAAGAGCGACAUAUGGUCCUCUUCCGAGCAAUUUGAGUA ((((((..(((.(((((((((...........((((....))))...........)))))))))..))).))))))(((..((((((.(((.....)))))))))..))).......... ( -41.15, z-score = -2.92, R) >droSec1.super_10 2609711 117 + 3036183 CUUUUUUCGCACCCACAAUUCUUCCCGUUUUGGUAGAAAUCU---GUCUUUCUUCGGAUUGUGGAAUGCCAGAAGGGCUGUGAAGAGCGACAUAUGGUCCUCUUCAGAGCAAUUUGAGUA ((((((..(((.((((((((((.((......)).)))).(((---(........))))))))))..))).))))))(((.(((((((.(((.....)))))))))).))).......... ( -38.20, z-score = -2.58, R) >droYak2.chrX 16276428 119 - 21770863 CCUUCCUCGCACCCACAAUUCUUCUCAUUUUGGUAGAAAUCUGCAGUCUUUCUUCGGAUUGUGGACUGCCAGAAGGGCUGUGAAGAGCGACAUAUGGUCCUCUGCAG-GCAAUUUGAGUA ....(((.((...((((.(((((((......(((((...((..((((((......))))))..)))))))))))))).)))).((((.(((.....)))))))))))-)........... ( -37.70, z-score = -1.44, R) >droEre2.scaffold_4690 293832 120 + 18748788 CCCUCUUCGCACCCACAAUUCUUCCCGUUUUGGUAGAAAUCUGCAGUCUUUCUUCGGUUUGUGGAAUGCCAGAAGGGCUGUGAAGAGCGACAUAUGGUCCUCUUCCGACCAAUUUGAGUA .(((((((((((((....((((.((......)).)))).((((((.(((..(....).....))).)).)))).))).))))))))).).((..(((((.......)))))...)).... ( -36.30, z-score = -1.85, R) >consensus CCUUCUUCGCACCCACAAUUCUUCCCGUUUUGGUAGAAAUCUGCAGUCUUUCUUCGGAUUGUGGAAUGCCAGAAGGGCUGUGAAGAGCGACAUAUGGUCCUCUUCAGAGCAAUUUGAGUA ((((((..(((.(((((((((...........((((....))))...........)))))))))..))).))))))(((.(((((((.(((.....)))))))))).))).......... (-33.95 = -36.03 + 2.08)

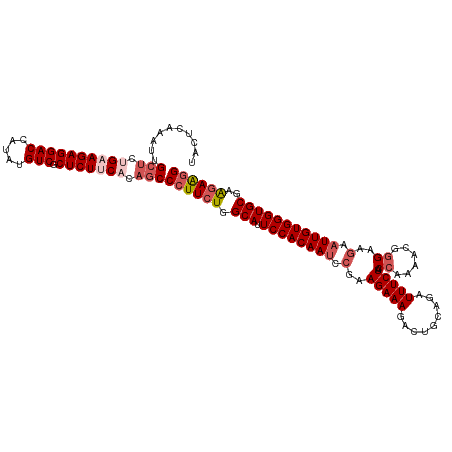
| Location | 2,868,485 – 2,868,605 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.25 |
| Shannon entropy | 0.10374 |
| G+C content | 0.46969 |
| Mean single sequence MFE | -35.03 |
| Consensus MFE | -30.94 |
| Energy contribution | -32.82 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.760386 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 2868485 120 - 22422827 UACUCAAAUUGCUCUGUAGAGGACCAUAUGUCGCUCUUCACAGCCCUUCUGGCAUUCCACAAUCGGAAGAAAGACUGCAGAUUUCUACCAAAACGGGAAGAAUUGUGGGUGCGAAGAAGG ..........(((.((.(((((((.....))).)))).)).)))((((((.((((.((((((((....))............((((.((......)).))))))))))))))..)))))) ( -38.50, z-score = -2.57, R) >droSim1.chrX 2010902 120 - 17042790 UACUCAAAUUGCUCGGAAGAGGACCAUAUGUCGCUCUUCACAGCCCUUCUGGCAUUCCACAAUCCGAAGAAAGACUGCAGAUUUCUACCAAAACGGGAAGAAUUGUGGGUGCGAAGAAGG ..........(((..(((((((((.....))).))))))..)))((((((.((((.(((((((.(..(((((.........))))).((.....))...).)))))))))))..)))))) ( -38.00, z-score = -2.50, R) >droSec1.super_10 2609711 117 - 3036183 UACUCAAAUUGCUCUGAAGAGGACCAUAUGUCGCUCUUCACAGCCCUUCUGGCAUUCCACAAUCCGAAGAAAGAC---AGAUUUCUACCAAAACGGGAAGAAUUGUGGGUGCGAAAAAAG ...((.....(((.((((((((((.....))).))))))).))).......((((.(((((((.(..(((((...---...))))).((.....))...).)))))))))))))...... ( -33.80, z-score = -2.54, R) >droYak2.chrX 16276428 119 + 21770863 UACUCAAAUUGC-CUGCAGAGGACCAUAUGUCGCUCUUCACAGCCCUUCUGGCAGUCCACAAUCCGAAGAAAGACUGCAGAUUUCUACCAAAAUGAGAAGAAUUGUGGGUGCGAGGAAGG ...........(-(((((((((((.....))).)))(((((((..(((((.((((((...............))))))((....)).........)))))..)))))))))).))).... ( -29.76, z-score = 0.08, R) >droEre2.scaffold_4690 293832 120 - 18748788 UACUCAAAUUGGUCGGAAGAGGACCAUAUGUCGCUCUUCACAGCCCUUCUGGCAUUCCACAAACCGAAGAAAGACUGCAGAUUUCUACCAAAACGGGAAGAAUUGUGGGUGCGAAGAGGG ..........(((.((((((((((.....))).)))))).).)))(((((.((((.((((((.....(((((.........))))).((.....))......))))))))))..))))). ( -35.10, z-score = -1.24, R) >consensus UACUCAAAUUGCUCUGAAGAGGACCAUAUGUCGCUCUUCACAGCCCUUCUGGCAUUCCACAAUCCGAAGAAAGACUGCAGAUUUCUACCAAAACGGGAAGAAUUGUGGGUGCGAAGAAGG ..........(((.((((((((((.....))).))))))).)))((((((.(((.((((((((.(..(((((.........))))).((......))..).)))))))))))..)))))) (-30.94 = -32.82 + 1.88)

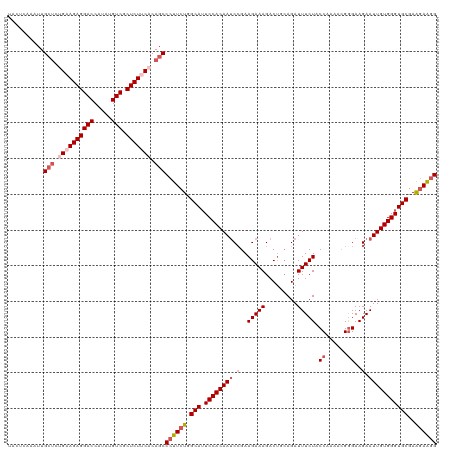
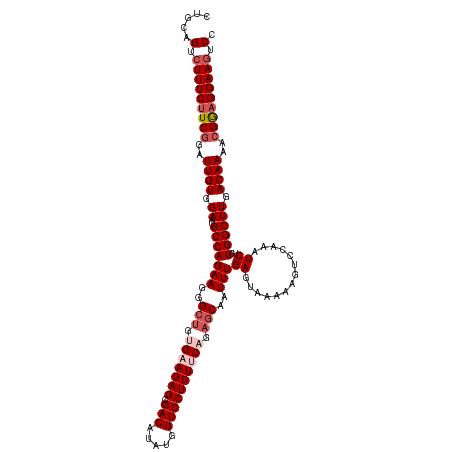
| Location | 2,868,525 – 2,868,645 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.58 |
| Shannon entropy | 0.13468 |
| G+C content | 0.46470 |
| Mean single sequence MFE | -41.68 |
| Consensus MFE | -32.20 |
| Energy contribution | -33.76 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.13 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.980831 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 2868525 120 + 22422827 CUGCAGUCUUUCUUCCGAUUGUGGAAUGCCAGAAGGGCUGUGAAGAGCGACAUAUGGUCCUCUACAGAGCAAUUUGAGUAAAAAGUCCAAAUCUGUGGCUUGACAAAACGGCGGAAAUCC (..(((((........)))))..)..((((.(((..(((.((.((((.(((.....))))))).)).)))..)))..((...(((((((....)).))))).)).....))))....... ( -35.90, z-score = -1.58, R) >droSim1.chrX 2010942 120 + 17042790 CUGCAGUCUUUCUUCGGAUUGUGGAAUGCCAGAAGGGCUGUGAAGAGCGACAUAUGGUCCUCUUCCGAGCAAUUUGAGUAAAAAGACCAAAUCUGUGGCUUGACAAAACGGAGGAAGUCC .....(.(((((((((..((((.((..((((..((((((..((((((.(((.....)))))))))..)))..((((.((......))))))))).)))))).))))..))))))))).). ( -40.60, z-score = -2.55, R) >droSec1.super_10 2609751 117 + 3036183 ---CUGUCUUUCUUCGGAUUGUGGAAUGCCAGAAGGGCUGUGAAGAGCGACAUAUGGUCCUCUUCAGAGCAAUUUGAGUAAAAAGUUCAAAUCUGUGGCUUGACAAAACGGAGGAAGUCC ---..(.(((((((((..((((.((..((((..((((((.(((((((.(((.....)))))))))).)))..((((((.......))))))))).)))))).))))..))))))))).). ( -45.50, z-score = -4.52, R) >droYak2.chrX 16276468 119 - 21770863 CUGCAGUCUUUCUUCGGAUUGUGGACUGCCAGAAGGGCUGUGAAGAGCGACAUAUGGUCCUCUGCAG-GCAAUUUGAGUAAAAAGUCUAAAUCUUUGGCUUGACAAAACGAAGGAAGUCC .....(.(((((((((((((.((((((((((((..(.((((..((((.(((.....)))))))))))-.)..)))).))....))))))))))((((......)))).))))))))).). ( -38.20, z-score = -1.96, R) >droEre2.scaffold_4690 293872 120 + 18748788 CUGCAGUCUUUCUUCGGUUUGUGGAAUGCCAGAAGGGCUGUGAAGAGCGACAUAUGGUCCUCUUCCGACCAAUUUGAGUAAAAACUCGAAAUCUCUGGCUUGACAAAACGAAGGAAGUCC .....(.(((((((((.(((((.((..((((((..((.((.((((((.(((.....))))))))))).))..(((((((....)))))))...)))))))).))))).))))))))).). ( -48.20, z-score = -5.02, R) >consensus CUGCAGUCUUUCUUCGGAUUGUGGAAUGCCAGAAGGGCUGUGAAGAGCGACAUAUGGUCCUCUUCAGAGCAAUUUGAGUAAAAAGUCCAAAUCUGUGGCUUGACAAAACGGAGGAAGUCC .....(.(((((((((..((((.((..(((((((..(((.(((((((.(((.....)))))))))).)))..)))((..............))..)))))).))))..))))))))).). (-32.20 = -33.76 + 1.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:10:25 2011