
| Sequence ID | dm3.chrX |
|---|---|
| Location | 2,858,464 – 2,858,556 |
| Length | 92 |
| Max. P | 0.974541 |

| Location | 2,858,464 – 2,858,556 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 54.12 |
| Shannon entropy | 0.62301 |
| G+C content | 0.41062 |
| Mean single sequence MFE | -14.40 |
| Consensus MFE | -9.63 |
| Energy contribution | -10.20 |
| Covariance contribution | 0.57 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.06 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.91 |
| SVM RNA-class probability | 0.974541 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
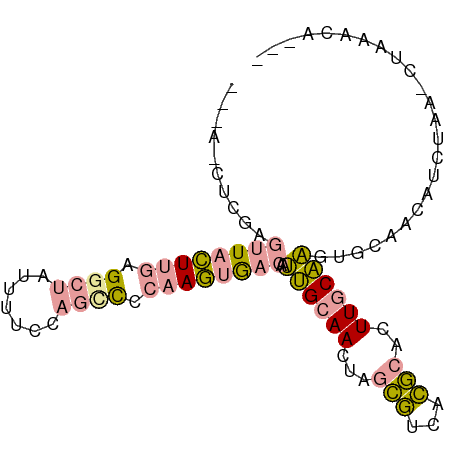
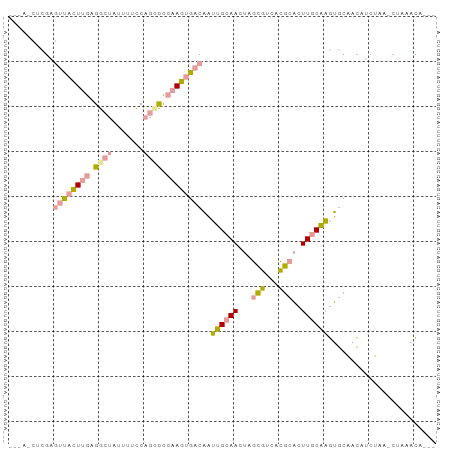
>dm3.chrX 2858464 92 + 22422827 --UAACUCGAGUUCCUUGAGGCUAUUUUCCAGCCCCAAGUGACAAUUGCAACUAGCGUCACGCACUUGCAAGUGCAACAUCUAA-CUAAACACAA --......((((..((((.((((.......)))).))))..))....((.....)).))..(((((....))))).........-.......... ( -20.80, z-score = -1.88, R) >droYak2.chrX 16265777 91 - 21770863 CCAACCUCGAGUUACUAGAGGCUACUUUCCAGUCCCAAGUGACAAUUGCAACUAGCGUCACGCACUUGCAAGUGCAACAUCUAA-CUAAACA--- ........((((((((.(.((((.......)))).).))))))....((.....)).))..(((((....))))).........-.......--- ( -17.60, z-score = -0.72, R) >dp4.chrXL_group1e 2311986 77 + 12523060 ------------UAAUUAAGAAGAUUAUUC--CUUCAAUUAUCCCGAGAAACCAAUAAAAUACCGUUCCUUAGACGAUACCCAAUCCAUAU---- ------------.......((((.......--))))...........................((((.....))))...............---- ( -4.80, z-score = -0.59, R) >consensus ___A_CUCGAGUUACUUGAGGCUAUUUUCCAGCCCCAAGUGACAAUUGCAACUAGCGUCACGCACUUGCAAGUGCAACAUCUAA_CUAAACA___ ..........((((((((.((((.......)))).))))))))..((((((...(((...)))..))))))........................ ( -9.63 = -10.20 + 0.57)

| Location | 2,858,464 – 2,858,556 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 54.12 |
| Shannon entropy | 0.62301 |
| G+C content | 0.41062 |
| Mean single sequence MFE | -23.50 |
| Consensus MFE | -13.88 |
| Energy contribution | -12.23 |
| Covariance contribution | -1.65 |
| Combinations/Pair | 1.52 |
| Mean z-score | -0.98 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.929780 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
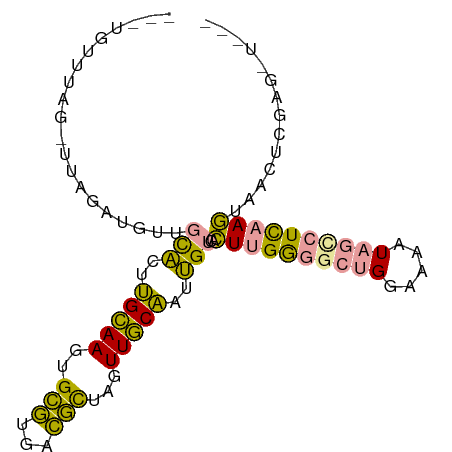
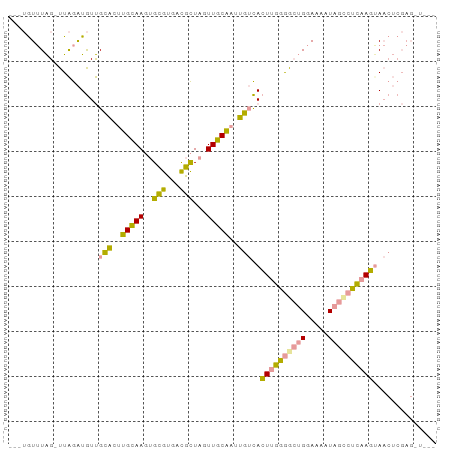
>dm3.chrX 2858464 92 - 22422827 UUGUGUUUAG-UUAGAUGUUGCACUUGCAAGUGCGUGACGCUAGUUGCAAUUGUCACUUGGGGCUGGAAAAUAGCCUCAAGGAACUCGAGUUA-- (((.((((..-...(((...(((((....)))))(..((....))..)....))).((((((((((.....)))))))))))))).)))....-- ( -31.70, z-score = -2.52, R) >droYak2.chrX 16265777 91 + 21770863 ---UGUUUAG-UUAGAUGUUGCACUUGCAAGUGCGUGACGCUAGUUGCAAUUGUCACUUGGGACUGGAAAGUAGCCUCUAGUAACUCGAGGUUGG ---..(((((-((....((.(((.((((((.((((...))).).)))))).))).))....)))))))...(((((((.((...)).))))))). ( -27.00, z-score = -0.92, R) >dp4.chrXL_group1e 2311986 77 - 12523060 ----AUAUGGAUUGGGUAUCGUCUAAGGAACGGUAUUUUAUUGGUUUCUCGGGAUAAUUGAAG--GAAUAAUCUUCUUAAUUA------------ ----..((.(((..(((((((((....).))))))))..))).)).........(((((((((--((....)))).)))))))------------ ( -11.80, z-score = 0.50, R) >consensus ___UGUUUAG_UUAGAUGUUGCACUUGCAAGUGCGUGACGCUAGUUGCAAUUGUCACUUGGGGCUGGAAAAUAGCCUCAAGUAACUCGAG_U___ ....................(((..(((((..(((...)))...)))))..)))..((((((((((.....)))))))))).............. (-13.88 = -12.23 + -1.65)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:10:22 2011