| Sequence ID | dm3.chr2L |
|---|---|
| Location | 9,813,789 – 9,813,889 |
| Length | 100 |
| Max. P | 0.628501 |

| Location | 9,813,789 – 9,813,889 |
|---|---|
| Length | 100 |
| Sequences | 9 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 68.15 |
| Shannon entropy | 0.65107 |
| G+C content | 0.30471 |
| Mean single sequence MFE | -11.81 |
| Consensus MFE | -7.06 |
| Energy contribution | -7.00 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.25 |
| Mean z-score | -0.28 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.628501 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
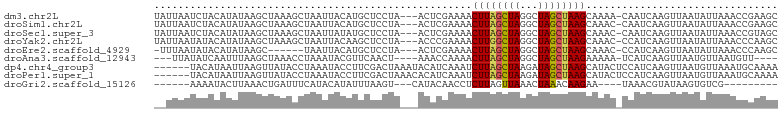
>dm3.chr2L 9813789 100 - 23011544 UAUUAAUCUACAUAUAAGCUAAAGCUAAUUACAUGCUCCUA---ACUCGAAAACUUAGCUAGGCUAGCUAAGCAAAA-CAAUCAAGUUAAUAUUAAACCGAAGC (((((((.........(((....)))...............---.........((((((((...)))))))).....-.......)))))))............ ( -12.30, z-score = 0.49, R) >droSim1.chr2L 9595294 100 - 22036055 UAUUAAUCUACAUAUAAGCUAAAGCUAAUUACAUGCUCCUA---ACUCGAAAACUUAGCUAGGCUAGCUAAGCAAAC-CAAUCAAGUUAAUAUUAAACCGAAGC (((((((.........(((....)))...............---.........((((((((...)))))))).....-.......)))))))............ ( -12.30, z-score = 0.46, R) >droSec1.super_3 5260504 100 - 7220098 UAUUAAUCUACAUAUAAGCUAAAGCUAAUUAUAUGCUCCUA---ACUCGAAAACUUAGCUAGGCUAGCUAAGCAAAC-CAAUCAAGUUAAUAUUAAACCGUAGC .......((((((((((((....)))...))))).....((---(((.((...((((((((...)))))))).....-...)).)))))..........)))). ( -15.60, z-score = -0.36, R) >droYak2.chr2L 12500353 100 - 22324452 UAUUAAUAUACAUAUAAGCUAAAGCUAAUUACAAGCUCCUA---ACCCGAAAACUUGGCUAGGCUAGCUAAGCAAAC-CCAUCAAGUUAAUAUUAAACCCAAGC ..(((((((.......(((....))).......((((((((---..((((....)))).))))..))))........-...........)))))))........ ( -14.60, z-score = -0.28, R) >droEre2.scaffold_4929 10425996 93 - 26641161 -UUUAAUAUACAUAUAAGC------UAAUUACAUGCUCCUA---ACUCGAAAACUUAGCUAGGCUAGCUAAGCAAAC-CCAUCAAGUUAAUAUUAAACCCAAGC -((((((((.......(((------.........)))..((---(((.((...((((((((...)))))))).....-...)).)))))))))))))....... ( -13.80, z-score = -1.08, R) >droAna3.scaffold_12943 1251936 92 - 5039921 ---UUAUAUCAAUUUAAGCUAAACCUAAAUACGUUCAACU----AAACCAAAACUUAGCUAGGCUAGCUAAGAAAAA-UCAUCAAGUUAAUGUUAAUGUU---- ---...........................(((((.((((----.........((((((((...)))))))).....-......))))))))).......---- ( -10.45, z-score = -0.36, R) >dp4.chr4_group3 11334094 98 + 11692001 ------UACAUAAUUAAGUUAUACCUAAAUACCUUCGACUAAAUACAUCAAAUCUUAGCUAAGAUAGCUAAGCAUACUCCAUCAAGUUAAUGUUAAAUGCAAAA ------..............................((((.............((((((((...))))))))............))))..(((.....)))... ( -8.81, z-score = -0.09, R) >droPer1.super_1 8449722 98 + 10282868 ------UACAUAAUUAAGUUAUACCUAAAUACCUUCGACUAAACACAUCAAAUCUUAGCUAAGAUAGCUAAGCAUACUCCAUCAAGUUAAUGUUAAAUGCAAAA ------..............................((((.............((((((((...))))))))............))))..(((.....)))... ( -8.81, z-score = -0.18, R) >droGri2.scaffold_15126 6739100 82 - 8399593 ------AAAAUACUUAAACUGAUUUCAUACAUAUUUAAGU---CAUACAACCUCUUAGUUAAACUAAACAAGAA----UAAACGUAUAAGUGUCG--------- ------...(((((((.((((((((.(.......).))))---)).......((((.(((......))))))).----.....)).)))))))..--------- ( -9.60, z-score = -1.15, R) >consensus __UUAAUAUACAUAUAAGCUAAAGCUAAUUACAUGCUCCUA___ACACGAAAACUUAGCUAGGCUAGCUAAGCAAAC_CCAUCAAGUUAAUAUUAAACCCAAGC .....................................................((((((((...))))))))................................ ( -7.06 = -7.00 + -0.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:29:01 2011