| Sequence ID | dm3.chrX |
|---|---|
| Location | 2,836,032 – 2,836,129 |
| Length | 97 |
| Max. P | 0.688076 |

| Location | 2,836,032 – 2,836,129 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 61.70 |
| Shannon entropy | 0.68046 |
| G+C content | 0.70365 |
| Mean single sequence MFE | -43.50 |
| Consensus MFE | -17.46 |
| Energy contribution | -18.26 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.61 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.688076 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
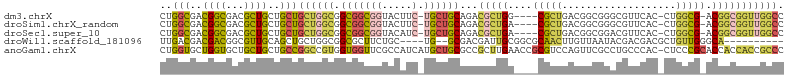
>dm3.chrX 2836032 97 - 22422827 CUGGCGACGGCGACGCUGCUGCUGCUGGCGGCGGCGGUACUUC-UGCUGCAGACGCUGG----CGCUGACGGCGGGCGUUCAC-CUGGCG-ACGGCGGUUGGCC ..(((..(((((.(((((((((.....))))))))).....((-((...))))))))).----(((((.((.((((......)-))).))-.)))))....))) ( -47.60, z-score = -0.58, R) >droSim1.chrX_random 1419246 97 - 5698898 CUGGCGACGGCGACGCUGCUGCUGCUGGCGGCGGCGGUACUUC-UGCUGCAGACGCUGA----CGCUGACGGCGGGCGUUCAC-CUGGCG-ACGGCGGUUGGCC ..(((..(((((.(((((((((.....))))))))).....((-((...))))))))).----(((((.((.((((......)-))).))-.)))))....))) ( -47.70, z-score = -1.22, R) >droSec1.super_10 2576940 97 - 3036183 CUGGCGACGGCGACGCUGCUGCUGCUGGCGGCGGCGGUACAUC-UGCUGCAGACGCUGA----CGCUGACGGCGGACGUUCAC-CUGGCG-ACGGCGGUUGGCC ..(((..((.((.(((((((((((....)))))))(((((.((-((((((((.((....----))))).))))))).))..))-).))))-.)).))....))) ( -45.30, z-score = -1.07, R) >droWil1.scaffold_181096 2231577 88 - 12416693 UUGACGACGACGGCGUUGCAGCUGCUGGCGGCGCUUCUGC----UG--GCGACGAUUGCGGCGCAACUUGUUAAUACGACGACGCUGUUGGGCA---------- .......((((((((((((.(((((...((.((((.....----.)--))).))...))))))))))(((((.....))))).)))))))....---------- ( -36.10, z-score = -1.72, R) >anoGam1.chrX 15077570 103 + 22145176 CUGGUGCUGGUGCUGCUGCUGCCGGCCGUGGUGGUUCGCCAUCAUGCUGCGCCGCUUGAACCGCGUCCAGUUCGCCUGCCCAC-CUCCCGCACCACCACCGCCC ..((((.((((((....((.((((((.(((((((....))))))))))).)).))..((((........))))..........-.....)))))).)))).... ( -40.80, z-score = -1.10, R) >consensus CUGGCGACGGCGACGCUGCUGCUGCUGGCGGCGGCGGUACAUC_UGCUGCAGACGCUGA____CGCUGACGGCGGACGUUCAC_CUGGCG_ACGGCGGUUGGCC ..(((..((((...))))..)))((((((.((((((........))))))...(((((.....(((.....)))..((((......))))..))))))))))). (-17.46 = -18.26 + 0.80)

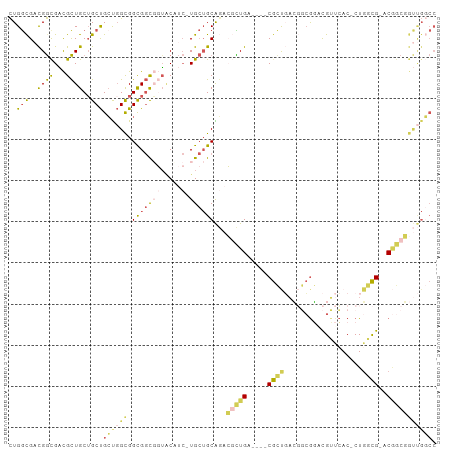
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:10:19 2011