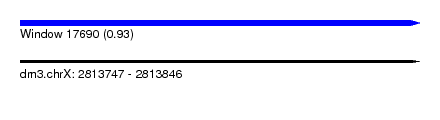
| Sequence ID | dm3.chrX |
|---|---|
| Location | 2,813,747 – 2,813,846 |
| Length | 99 |
| Max. P | 0.932184 |

| Location | 2,813,747 – 2,813,846 |
|---|---|
| Length | 99 |
| Sequences | 9 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 73.47 |
| Shannon entropy | 0.55006 |
| G+C content | 0.45932 |
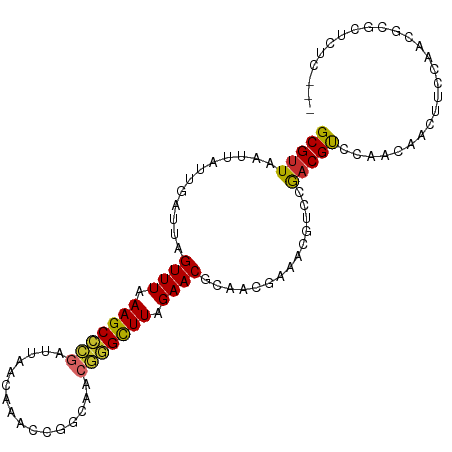
| Mean single sequence MFE | -22.15 |
| Consensus MFE | -14.42 |
| Energy contribution | -13.92 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.932184 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
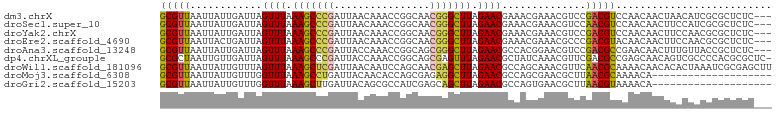
>dm3.chrX 2813747 99 + 22422827 GCGUUAAUUAUUGAUUAGUUUAAAGCCCGAUUAACAAACCGGCAACGGGCUUAGAACGAAACGAAACGUCCGACGUCCAACAACUAACAUCGCGCUCUC--- ((((......(((....((((.(((((((.((..(.....)..))))))))).))))....))).(((.....)))...............))))....--- ( -20.90, z-score = -0.85, R) >droSec1.super_10 2554889 99 + 3036183 GCGUUAAUUAUUGAUUAGUUUAAAGCCCGAUUAACAAACCGGCAACGGGCUUAGAACGAAACGAAACGUCCAACGUCCAACAACUUCCAUCGCGCUCUC--- ((((......(((....((((.(((((((.((..(.....)..))))))))).))))....))).(((.....)))...............))))....--- ( -20.90, z-score = -1.15, R) >droYak2.chrX 16225244 99 - 21770863 GCGUUAAUUAUUGAUUAGUUUAAAGCCCGAUUAACAAACCGGCAACGGGCUUAGAACGAAACGAAACGUCCGACGUCCAACAACUUCCAACGCGCUCUC--- (((((.....(((....((((.(((((((.((..(.....)..))))))))).))))........(((.....)))....))).....)))))......--- ( -22.60, z-score = -1.30, R) >droEre2.scaffold_4690 236253 99 + 18748788 GCGUUAAUUACUGAUUAGUUUAAAGCCCGAUUAACAAACCGGCAACGGGCUUAGAACGAAACGAAACGCCCGACGUACAACAACUUCCAACGCGCUCUC--- (((((............((((.(((((((.((..(.....)..))))))))).))))........(((.....)))............)))))......--- ( -22.20, z-score = -1.43, R) >droAna3.scaffold_13248 4671529 99 - 4840945 GCGUUAAUUAUUGAUUAGUUUAAAGCCCGAUUACCAAACCGGCAGCGGGCUUAGAACGCCACGGAACGUCCGACGCCGAACAACUUUGUUACCGCUCUC--- (((.((((..(((....((((.(((((((....((.....))...))))))).))))((..(((.....)))..))....)))....)))).)))....--- ( -27.90, z-score = -1.73, R) >dp4.chrXL_group1e 2260432 101 + 12523060 GCGCUAAUUGUUGAUUAGUUUAAAGCCCGAUUACCAAACCGGCAGCGAGUUUAGAACGCUAUCAAACGUUCGACGCCGAGCAACAGUCGCCCCACGCGCUC- ((((......(((((..((((.((((.((....((.....))...)).)))).))))...)))))..(((((....)))))..............))))..- ( -25.30, z-score = -0.76, R) >droWil1.scaffold_181096 2201781 102 + 12416693 GCGUUAAUUAUUGUUUAGUUUAAAGCUCGAUUAACAAUCCAGCAACGAGCUUAGAACGCCAGCAAACGUUCAACGCAAAACAACACACUAAAUCGCGAGCUU ((((((((..(((((..((((.(((((((................))))))).))))...)))))..))).))))).......................... ( -21.29, z-score = -1.67, R) >droMoj3.scaffold_6308 1759912 82 + 3356042 GCGUUAAUUAUUGUUUGGUUUAAAGCCUGAUUACAACACCAGCGAGAGGCUUAGAACGCCAGCGAACGCUUAACGCAAAACA-------------------- (((((((...(((((.(((((.((((((..(..(.......)..).)))))).)))).).)))))....)))))))......-------------------- ( -21.60, z-score = -1.91, R) >droGri2.scaffold_15203 2333458 82 + 11997470 GCGUUAAUUAUUGUUUGGUUUAAAGCUUGAUUACAGCGCCAUCGAGCAGCUUAGAACGCCAGUGAACGCUUAACGUAAAACA-------------------- (((((.....(((((((((.....(((.......)))...))))))))).....)))))..((..(((.....)))...)).-------------------- ( -16.70, z-score = 0.41, R) >consensus GCGUUAAUUAUUGAUUAGUUUAAAGCCCGAUUAACAAACCGGCAACGGGCUUAGAACGCAACGAAACGUCCGACGUCCAACAACUUCCAACGCGCUCUC___ (((((............((((.(((((((................))))))).))))..............))))).......................... (-14.42 = -13.92 + -0.50)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:10:18 2011