
| Sequence ID | dm3.chrX |
|---|---|
| Location | 2,790,299 – 2,790,404 |
| Length | 105 |
| Max. P | 0.607450 |

| Location | 2,790,299 – 2,790,404 |
|---|---|
| Length | 105 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 90.91 |
| Shannon entropy | 0.17820 |
| G+C content | 0.41879 |
| Mean single sequence MFE | -25.02 |
| Consensus MFE | -17.75 |
| Energy contribution | -17.48 |
| Covariance contribution | -0.26 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.607450 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
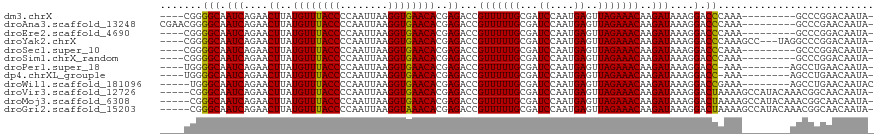
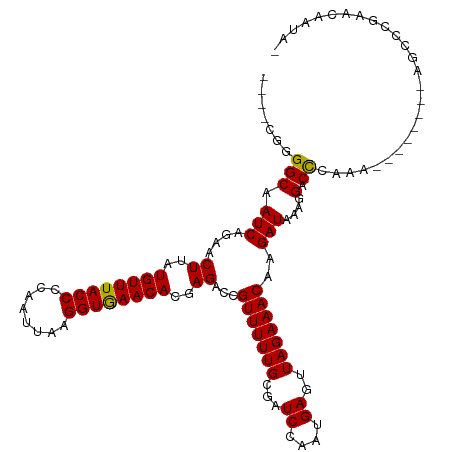
>dm3.chrX 2790299 105 - 22422827 ----CGGGGCAAUCAGAACUUAUGUUUACCCCAAUUAAGGUGAACACGAGACCGUUUUUGCGAUCCAAUGAGUUAGAAACAAGAUAAAGGACCCAAA---------GCCCGGACAAUA- ----(.((((.(((....((..((((((((........))))))))..))...(((((((...((....))..)))))))..)))...(....)...---------)))).)......- ( -26.60, z-score = -2.38, R) >droAna3.scaffold_13248 4637660 109 + 4840945 CGAACGGGGCAAUCAGAACUUAUGUUUACCCCAAUUAAGGUGAACACGAGACCGUUUUUGCGAUCCAAUGAGUUAGAAACAAGAUAAAGGACCCAAA---------GCCCGAACAAUA- ......((((.(((....((..((((((((........))))))))..))...(((((((...((....))..)))))))..)))...(....)...---------))))........- ( -25.80, z-score = -2.46, R) >droEre2.scaffold_4690 212190 105 - 18748788 ----CGGGGCAAUCAGAACUUAUGUUUACCCCAAUUAAGGUGAACACGAGACCGUUUUUGCGAUCCAAUGAGUUAGAAACAAGAUAAAGGACCCAAA---------GCCCGGACAAUA- ----(.((((.(((....((..((((((((........))))))))..))...(((((((...((....))..)))))))..)))...(....)...---------)))).)......- ( -26.60, z-score = -2.38, R) >droYak2.chrX 16201020 111 + 21770863 ----CGGGGCAAUCAGAACUUAUGUUUACCCCAAUUAAGGUGAACACGAGACCGUUUUUGCGAUCCAAUGAGUUAGAAACAAGAUAAAGGACCCAAAGCC---UAGGCCCGGACAAUA- ----((((.(.(((....((..((((((((........))))))))..))...(((((((...((....))..)))))))..)))..(((........))---).).)))).......- ( -28.90, z-score = -2.15, R) >droSec1.super_10 2531566 105 - 3036183 ----CGGGGCAAUCAGAACUUAUGUUUACCCCAAUUAAGGUGAACACGAGACCGUUUUUGCGAUCCAAUGAGUUAGAAACAAGAUAAAGGACCCAAA---------GCCCGGACAAUA- ----(.((((.(((....((..((((((((........))))))))..))...(((((((...((....))..)))))))..)))...(....)...---------)))).)......- ( -26.60, z-score = -2.38, R) >droSim1.chrX_random 1407461 105 - 5698898 ----CGGGGCAAUCAGAACUUAUGUUUACCCCAAUUAAGGUGAACACGAGACCGUUUUUGCGAUCCAAUGAGUUAGAAACAAGAUAAAGGACCCAAA---------GCCCGGACAAUA- ----(.((((.(((....((..((((((((........))))))))..))...(((((((...((....))..)))))))..)))...(....)...---------)))).)......- ( -26.60, z-score = -2.38, R) >droPer1.super_18 1103640 105 - 1952607 ----UGGGGCAAUCAGAACUUAUGUUUACCCCAAUUAAGGUGAACACGAGACCGUUUUUGCGAUCCAAUGAGUUAGAAACAAGAUAAAGGACC-AAA--------AGCCUGAACAAUA- ----........((((..((..((((((((........))))))))..))...(((((((.(.(((......................)))))-)))--------)))))))......- ( -23.15, z-score = -1.71, R) >dp4.chrXL_group1e 2232737 105 - 12523060 ----UGGGGCAAUCAGAACUUAUGUUUACCCCAAUUAAGGUGAACACGAGACCGUUUUUGCGAUCCAAUGAGUUAGAAACAAGAUAAAGGACC-AAA--------AGCCUGAACAAUA- ----........((((..((..((((((((........))))))))..))...(((((((.(.(((......................)))))-)))--------)))))))......- ( -23.15, z-score = -1.71, R) >droWil1.scaffold_181096 2168657 106 - 12416693 -----UGGGCAAUCAGAACUUAUGUUUACCCCAAUUAAGGUGAACACGAGACCGUUUUUGCGAUCCAAUGAGUUAGAAACAAGAUAAAGGACCGAAA--------AGCCUGAACAAUAC -----.......((((..((..((((((((........))))))))..))...((((((.((.(((......................))).)))))--------)))))))....... ( -23.05, z-score = -2.18, R) >droVir3.scaffold_12726 91845 113 - 2840439 -----CGGGCAAUCAGAACUUAUGUUUACCCCAAUUAAGGUGAACACGAGACCGUUUUUGCGAUCCAAUGAGUUAGAAACAAGAUAAAGGACUAAAAGCCAUACAAACGGCAACAAUA- -----..(((........((..((((((((........))))))))..))...(((((((...((....))..))))))).................)))........(....)....- ( -23.30, z-score = -1.77, R) >droMoj3.scaffold_6308 1716633 113 - 3356042 -----CGGGCAAUCAGAACUUAUGUUUACCCCAAUUAAGGUGAACACGAGACCGUUUUUGCGAUCCAAUGAGUUAGAAACAAGAUAAAGGACUAAAAGCCAUACAAACGGCAACAAUA- -----..(((........((..((((((((........))))))))..))...(((((((...((....))..))))))).................)))........(....)....- ( -23.30, z-score = -1.77, R) >droGri2.scaffold_15203 2304557 113 - 11997470 -----CGGGCAAUCAGAACUUAUGUUUACCCCAAUUAAGGUAAACACGAGACCGUUUUUGCGAUCCAAUGAGUUAGAAACAAGAUAAAGGACUAAAAGCCAUACAAACGGCAACAAUA- -----..(((........((..((((((((........))))))))..))...(((((((...((....))..))))))).................)))........(....)....- ( -23.20, z-score = -2.05, R) >consensus ____CGGGGCAAUCAGAACUUAUGUUUACCCCAAUUAAGGUGAACACGAGACCGUUUUUGCGAUCCAAUGAGUUAGAAACAAGAUAAAGGACCCAAA________AGCCCGAACAAUA_ .......(((.(((....((..((((((((........))))))))..))...(((((((...((....))..)))))))..)))....).)).......................... (-17.75 = -17.48 + -0.26)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:10:16 2011