| Sequence ID | dm3.chrX |
|---|---|
| Location | 2,749,164 – 2,749,298 |
| Length | 134 |
| Max. P | 0.987598 |

| Location | 2,749,164 – 2,749,281 |
|---|---|
| Length | 117 |
| Sequences | 7 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 81.67 |
| Shannon entropy | 0.32097 |
| G+C content | 0.33916 |
| Mean single sequence MFE | -21.07 |
| Consensus MFE | -11.20 |
| Energy contribution | -10.33 |
| Covariance contribution | -0.87 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.890752 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
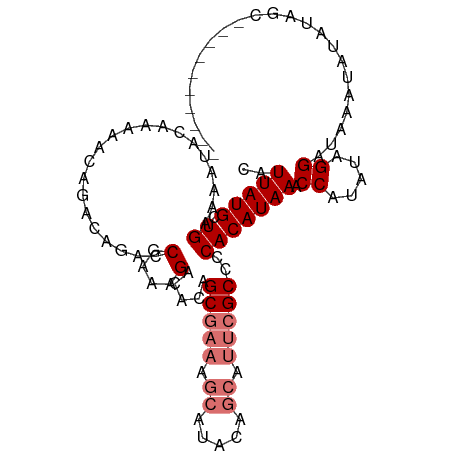
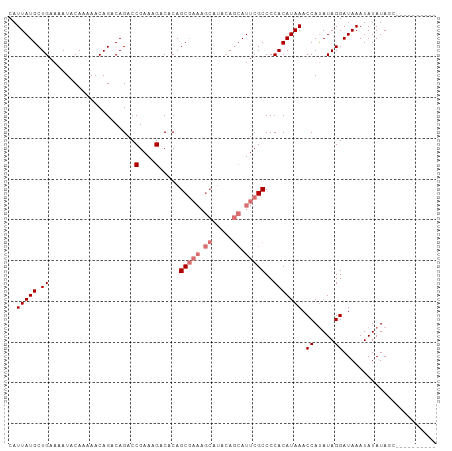
>dm3.chrX 2749164 117 + 22422827 UGUGUGUCUGGCAUAAAUACAGUUUACUAAAAUUGAAUUCAUUAUGCUGAAAAUACAAAAACAGACAGACCGAAAGACACAGCGAAAGCAUACAGCAUUCGCCCCACAUAAACCAUA ((((((((((((((((((.((((((....)))))).))...))))))((......))....)))))....(....))))))(((((.((.....)).)))))............... ( -26.70, z-score = -3.89, R) >droEre2.scaffold_4690 169495 104 + 18748788 UGUGUGCCUGGCAUAAAUACAGUUUACUAAAAUUGAAUUCAUUAUGCUGAAAAUACAAAAACAGACAGACCGAAAGACACAGC-------------AUUCGCCCCACAUAAACCAUA ((((((...(((.......((((((....))))))........((((((............(.....)..(....)...))))-------------))..))).))))))....... ( -19.00, z-score = -2.36, R) >droYak2.chrX 16159102 104 - 21770863 UAUGGGUCUGGCAUAAAUACAGUUUACUAAAAUUGAAUUCAUUAUGCUGAAAAUACAAAAACAGACAGACCGAAAGACACAGC-------------AUUCGCCCCACAUAAACCACA ((((((((((((((((((.((((((....)))))).))...))))))((......))........))))))..........((-------------....))....))))....... ( -18.10, z-score = -2.07, R) >droSec1.super_10 2491105 117 + 3036183 UGUGUGUCUGGCAUAAAUACAGUUUACUAAAAUUGAAUUCAUUAUGCUGAAAAUACAAAAACAGACAGACCGAAAGACACAGCGAAAGCAUACAGCAUUCGCCCCACAUAAACCAUA ((((((((((((((((((.((((((....)))))).))...))))))((......))....)))))....(....))))))(((((.((.....)).)))))............... ( -26.70, z-score = -3.89, R) >droSim1.chrX 1908092 117 + 17042790 UGUGUGUCUGGCAUAAAUACAGUUUACUAAAAUUGAAUUCAUUAUGCUGAAAAUACAAAAACAGACAGACCGAAAGACACAGCGAAAGCAUACAGCAUUCGCCCCACAUAAACCAUA ((((((((((((((((((.((((((....)))))).))...))))))((......))....)))))....(....))))))(((((.((.....)).)))))............... ( -26.70, z-score = -3.89, R) >dp4.chrXL_group1e 2179865 98 + 12523060 UUUGUGUUUGGCAUAAAUGAAGUUUAGUAAAAUUCAAUUUAUUAUGCUAAAAAUACAAAAA---AAAGAGGGAACAUCAUCG----------------UCGUUUCACAGAAACCAUA ((((((((((((((((...(((((.(((...))).))))).))))))))..))))))))..---......(((((.......----------------..)))))............ ( -15.40, z-score = -1.06, R) >droPer1.super_18 1050247 98 + 1952607 UUUGUGUUUGGCAUAAAUGAAGUUUACUAAAAUUCAAUUUAUUAUGCUAAAAAUACAAAAA---AAAGAGGGAAUAUCAUCG----------------UCGUUUCACAGAAACCAUA ((((((((((((((((.((((................))))))))))))..))))))))..---...((.((.......)).----------------))(((((...))))).... ( -14.89, z-score = -0.97, R) >consensus UGUGUGUCUGGCAUAAAUACAGUUUACUAAAAUUGAAUUCAUUAUGCUGAAAAUACAAAAACAGACAGACCGAAAGACACAGC_____________AUUCGCCCCACAUAAACCAUA ((((((...(((.......((((((....))))))..((((......)))).................................................))).))))))....... (-11.20 = -10.33 + -0.87)
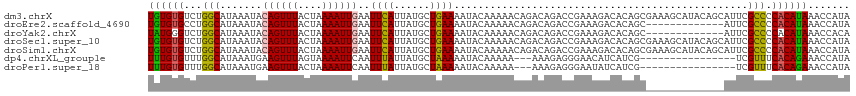
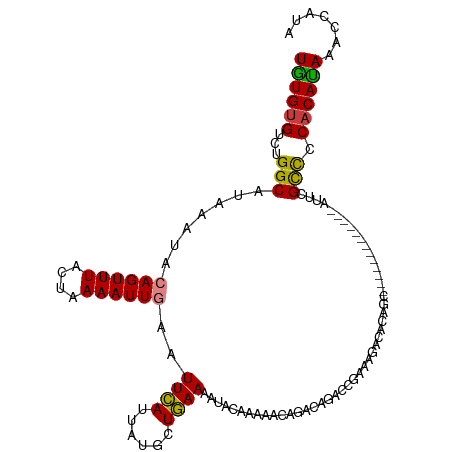
| Location | 2,749,203 – 2,749,298 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 86.83 |
| Shannon entropy | 0.20269 |
| G+C content | 0.36854 |
| Mean single sequence MFE | -13.00 |
| Consensus MFE | -9.73 |
| Energy contribution | -10.60 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.07 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.28 |
| SVM RNA-class probability | 0.987598 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 2749203 95 + 22422827 CAUUAUGCUGAAAAUACAAAAACAGACAGACCGAAAGACACAGCGAAAGCAUACAGCAUUCGCCCCACAUAAACCAUAUAGGAUAAAUAUAUAUC---------- ....((((((.....................(....).....((....))...))))))..............((.....)).............---------- ( -14.30, z-score = -3.82, R) >droYak2.chrX 16159141 92 - 21770863 CAUUAUGCUGAAAAUACAAAAACAGACAGACCGAAAGACACAGC-------------AUUCGCCCCACAUAAACCACAUAGGAUAAAUAUAUAUACAAGUGUAUA ....((((((............(.....)..(....)...))))-------------))..............((.....)).....(((((((....))))))) ( -9.10, z-score = -1.10, R) >droSec1.super_10 2491144 95 + 3036183 CAUUAUGCUGAAAAUACAAAAACAGACAGACCGAAAGACACAGCGAAAGCAUACAGCAUUCGCCCCACAUAAACCAUAUAGGAUAAAUAUAUAGC---------- ....((((((.....................(....).....((....))...))))))..............((.....)).............---------- ( -14.30, z-score = -3.68, R) >droSim1.chrX 1908131 95 + 17042790 CAUUAUGCUGAAAAUACAAAAACAGACAGACCGAAAGACACAGCGAAAGCAUACAGCAUUCGCCCCACAUAAACCAUAUAGGAUAAAUAUAUAGC---------- ....((((((.....................(....).....((....))...))))))..............((.....)).............---------- ( -14.30, z-score = -3.68, R) >consensus CAUUAUGCUGAAAAUACAAAAACAGACAGACCGAAAGACACAGCGAAAGCAUACAGCAUUCGCCCCACAUAAACCAUAUAGGAUAAAUAUAUAGC__________ ..(((((.((.....................(....).....(((((.((.....)).)))))..))))))).((.....))....................... ( -9.73 = -10.60 + 0.87)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:10:14 2011