| Sequence ID | dm3.chrX |
|---|---|
| Location | 2,733,469 – 2,733,583 |
| Length | 114 |
| Max. P | 0.659502 |

| Location | 2,733,469 – 2,733,583 |
|---|---|
| Length | 114 |
| Sequences | 7 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 64.80 |
| Shannon entropy | 0.65910 |
| G+C content | 0.41850 |
| Mean single sequence MFE | -27.40 |
| Consensus MFE | -11.12 |
| Energy contribution | -11.56 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.659502 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
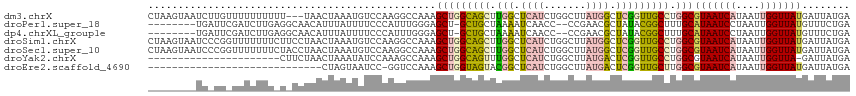
>dm3.chrX 2733469 114 - 22422827 CUAAGUAAUCUUGUUUUUUUUUU---UAACUAAAUGUCCAAGGCCAAAGCUGGCAGCUUGGCUCAUCUGGCUUAUGGCUCGGUUGCCUGGCGUAAUCAUAAUUGGUUAUGAUUAUGA .........((((..(.(((...---.....))).)..))))((((.....(((((((.(((.(((.......)))))).)))))))))))((((((((((....)))))))))).. ( -30.60, z-score = -1.32, R) >droPer1.super_18 1031568 106 - 1952607 --------UGAUUCGAUCUUGAGGCAACAUUUAUUUUCCCAUUUGGGAGCU-GCUGCUAAAAUCAACC--CCGAACGCUAUACGGCUUUGCAUAAUCCUAAUUGGUUAUGUUUCUGA --------...((((...((((((((.((......(((((....))))).)-).))))....))))..--.)))).......(((....((((((((......))))))))..))). ( -20.70, z-score = 0.05, R) >dp4.chrXL_group1e 2162296 106 - 12523060 --------UGAUUCGAUCUUGAGGCAACAUUUAUUUUCCCAUUUGGGAGCU-GCUGCUAAAAUCAACC--CCGAACGCUAUACGGCUUUGCAUAAUCCUAAUUGGUUAUGUUUCUGA --------...((((...((((((((.((......(((((....))))).)-).))))....))))..--.)))).......(((....((((((((......))))))))..))). ( -20.70, z-score = 0.05, R) >droSim1.chrX 1900029 117 - 17042790 CUAAGUAAUCCCGGUUUUUUUCUUCCUAACUAAAUGUCCAAGGCCAAAGCUGGCAGCUUGGCUCAUCUGGCUUAUGGCUCGGUUGCCUGGCGUAAUCAUAAUUGGUUAUGAUUAUGA (((.((((((((((((((..((((....((.....))..))))..)))))))).((((.((((.....))))...)))).)))))).)))(((((((((((....))))))))))). ( -31.80, z-score = -0.82, R) >droSec1.super_10 2483073 117 - 3036183 CUAAGUAAUCCCGGUUUUUUUCUACCUAACUAAAUGUCCAAGGCCAAAGCUGGCAGCUUGGCUCAUCUGGCUUAUGGCUCGGUUGCCUGGCGUAAUCAUAAUUGGUUAUGAUUAUGA (((.((((((..(((........)))...............(((((((((..(.(((...)))...)..)))).))))).)))))).)))(((((((((((....))))))))))). ( -33.50, z-score = -1.22, R) >droYak2.chrX 16150777 94 + 21770863 ----------------------CUUCUAACUAAAUAUCCAAAGCCAAAGCUGGCAGUUUGGCUCAUCUGGCUUAUGACUCGGUUGCCUGGCGUAAUCAUAAUUGGUUA-GAUUAUGA ----------------------..(((((((((.(((.....((((.(((((..((((.((((.....))))...)))))))))...))))......))).)))))))-))...... ( -25.20, z-score = -1.51, R) >droEre2.scaffold_4690 161239 87 - 18748788 -----------------------------CUAGUAAUCC-GGUCCAAAGCUGGUAGUACGGCUCAUCUGGCUUAUGACUCGGUUGCUUGGCGUAAUCAUAAUUGGUUAUGAUUAUGA -----------------------------(.(((((((.-((((..((((..(.((.....))...)..))))..)))).))))))).).(((((((((((....))))))))))). ( -29.30, z-score = -3.21, R) >consensus ________UC_UG_UUUUUUU_U_CCUAACUAAAUGUCCAAGGCCAAAGCUGGCAGCUUGGCUCAUCUGGCUUAUGGCUCGGUUGCCUGGCGUAAUCAUAAUUGGUUAUGAUUAUGA ................................................(((((((((.(((.((((.......)))).))))))))).)))(((((((....)))))))........ (-11.12 = -11.56 + 0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:10:12 2011