
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 9,808,558 – 9,808,653 |
| Length | 95 |
| Max. P | 0.788175 |

| Location | 9,808,558 – 9,808,653 |
|---|---|
| Length | 95 |
| Sequences | 7 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 73.23 |
| Shannon entropy | 0.49876 |
| G+C content | 0.43948 |
| Mean single sequence MFE | -23.40 |
| Consensus MFE | -13.34 |
| Energy contribution | -14.29 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.693021 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
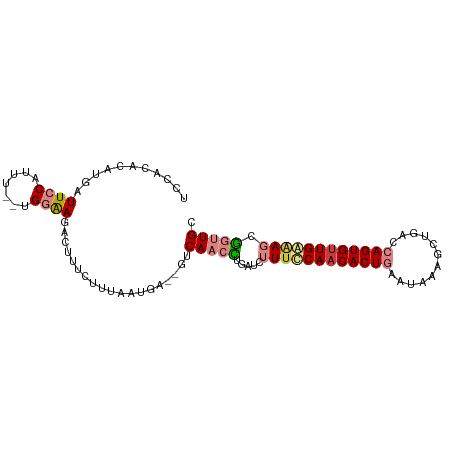
>dm3.chr2L 9808558 95 + 23011544 UCCACACAUGAUUCCAUUU--UGGAAGACUUUCUUUAACAAA-CUCAACCUGAUCUUUUCAACACUGCAUAAAGCUGACCAGUGUUGAGAGCGGUUGC ((((...(((....)))..--)))).................-..(((((.....((((((((((((((......))..)))))))))))).))))). ( -20.60, z-score = -1.23, R) >droSim1.chr2L 9590143 95 + 22036055 UCCACACAUGAUUCCAUUU--UGGAAGACUUUCCUUAAAUAA-CUCAACCGGAUCUUUUCAACACUGAAUAAAGCUGACCAGUGUUGAGAGCGGUUGC ((((...(((....)))..--)))).................-..((((((....((((((((((((............)))))))))))))))))). ( -24.20, z-score = -2.40, R) >droSec1.super_3 5255397 94 + 7220098 UCCACACAUGAUUCCAUUU--UGGAAGACUUUCUUUAAUAA--CUCAACCUGAUCUUAUCAACACUGAAUAAAGCUGACCAGUGUUGAGAGCGGUUGC ((((...(((....)))..--))))................--..(((((....(((.(((((((((............)))))))))))).))))). ( -20.30, z-score = -1.45, R) >droYak2.chr2L 12495117 94 + 22324452 UCCACACAUGAUUCCGUUU--UGGAAGACUUUUUAUAAGGA--GUCAACUGCGACUUUGCAACACUGCAUUAAGCUGGCCAGUGUUGAAAGCGGUUGC ((((...(((....)))..--)))).(((((((....))))--)))((((((.......(((((((((.........).))))))))...)))))).. ( -25.30, z-score = -1.00, R) >droEre2.scaffold_4929 10420924 94 + 26641161 UCCACACAUGAUUCCGUUU--UGGAAGACUUUUUAUAAGGA--GUCAACCGUGUCUUUCCAACACUGUAUAAAGCUGACCAGUGCUGAAAGCGGUUGC ((((...(((....)))..--)))).(((((((....))))--)))(((((...(((((...(((((..((....))..)))))..)))))))))).. ( -21.80, z-score = -0.61, R) >dp4.chr4_group3 11328323 94 - 11692001 UCCACACAUGAUUCCUCCUGUUGUGAGACAAACCUACGUGAGAGGCAAAUUUCCGCUUCCAACACUGGACGGUGUU----AGUGUUGGAAACAGCUGC ((((((((.((....)).)).)))).))....(((.(....)))).........((((((((((((((......))----)))))))))...)))... ( -25.80, z-score = -0.81, R) >droPer1.super_1 8443870 94 - 10282868 UCCACACAUGAUUCCUCCUGUUGUGAGACAAACCUACGUGAGAGGCAAAUUUCCGCUUCCAACACUGGACGGUGUU----AGUGUUGGAAACAGCUGC ((((((((.((....)).)).)))).))....(((.(....)))).........((((((((((((((......))----)))))))))...)))... ( -25.80, z-score = -0.81, R) >consensus UCCACACAUGAUUCCAUUU__UGGAAGACUUUCUUUAAUGA__GUCAACCUGAUCUUUCCAACACUGAAUAAAGCUGACCAGUGUUGAAAGCGGUUGC ...........((((.......))))...................(((((.....((((((((((((............)))))))))))).))))). (-13.34 = -14.29 + 0.94)

| Location | 9,808,558 – 9,808,653 |
|---|---|
| Length | 95 |
| Sequences | 7 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 73.23 |
| Shannon entropy | 0.49876 |
| G+C content | 0.43948 |
| Mean single sequence MFE | -26.84 |
| Consensus MFE | -13.67 |
| Energy contribution | -13.80 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.788175 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
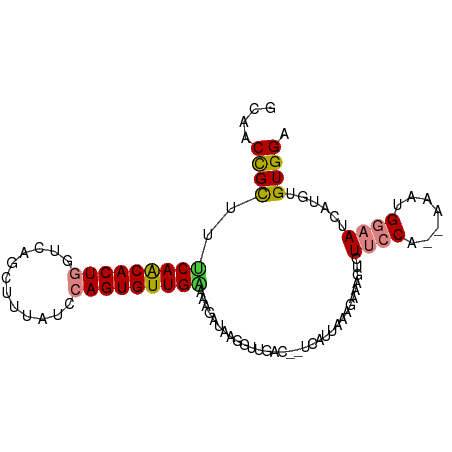
>dm3.chr2L 9808558 95 - 23011544 GCAACCGCUCUCAACACUGGUCAGCUUUAUGCAGUGUUGAAAAGAUCAGGUUGAG-UUUGUUAAAGAAAGUCUUCCA--AAAUGGAAUCAUGUGUGGA ....((((.((((((.((((((.((.....)).....(....)))))))))))))-................((((.--....))))......)))). ( -24.60, z-score = -1.07, R) >droSim1.chr2L 9590143 95 - 22036055 GCAACCGCUCUCAACACUGGUCAGCUUUAUUCAGUGUUGAAAAGAUCCGGUUGAG-UUAUUUAAGGAAAGUCUUCCA--AAAUGGAAUCAUGUGUGGA .((((((.((((((((((((..........))))))))))...))..))))))..-.................((((--..(((....)))...)))) ( -25.20, z-score = -1.56, R) >droSec1.super_3 5255397 94 - 7220098 GCAACCGCUCUCAACACUGGUCAGCUUUAUUCAGUGUUGAUAAGAUCAGGUUGAG--UUAUUAAAGAAAGUCUUCCA--AAAUGGAAUCAUGUGUGGA .(((((.((.((((((((((..........))))))))))..))....)))))..--................((((--..(((....)))...)))) ( -23.80, z-score = -1.40, R) >droYak2.chr2L 12495117 94 - 22324452 GCAACCGCUUUCAACACUGGCCAGCUUAAUGCAGUGUUGCAAAGUCGCAGUUGAC--UCCUUAUAAAAAGUCUUCCA--AAACGGAAUCAUGUGUGGA ....(((((((((((((((..((......)))))))))).)))))((((...(((--(..........))))((((.--....))))...)))).)). ( -22.60, z-score = -0.63, R) >droEre2.scaffold_4929 10420924 94 - 26641161 GCAACCGCUUUCAGCACUGGUCAGCUUUAUACAGUGUUGGAAAGACACGGUUGAC--UCCUUAUAAAAAGUCUUCCA--AAACGGAAUCAUGUGUGGA .((((((((((((((((((............))))))).)))))...))))))..--(((.((((.......((((.--....))))...)))).))) ( -25.10, z-score = -1.36, R) >dp4.chr4_group3 11328323 94 + 11692001 GCAGCUGUUUCCAACACU----AACACCGUCCAGUGUUGGAAGCGGAAAUUUGCCUCUCACGUAGGUUUGUCUCACAACAGGAGGAAUCAUGUGUGGA ((((((((((((((((((----..........))))))))))))))....)))).((.(((((.(((((.(((.......))).)))))))))).)). ( -33.30, z-score = -2.46, R) >droPer1.super_1 8443870 94 + 10282868 GCAGCUGUUUCCAACACU----AACACCGUCCAGUGUUGGAAGCGGAAAUUUGCCUCUCACGUAGGUUUGUCUCACAACAGGAGGAAUCAUGUGUGGA ((((((((((((((((((----..........))))))))))))))....)))).((.(((((.(((((.(((.......))).)))))))))).)). ( -33.30, z-score = -2.46, R) >consensus GCAACCGCUUUCAACACUGGUCAGCUUUAUCCAGUGUUGAAAAGAUAAGGUUGAC__UCAUUAAAGAAAGUCUUCCA__AAAUGGAAUCAUGUGUGGA ....((((..(((((((((............)))))))))................................((((.......))))......)))). (-13.67 = -13.80 + 0.13)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:29:00 2011