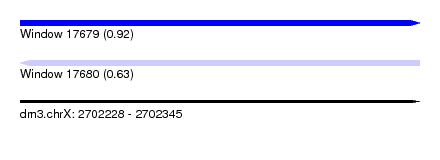
| Sequence ID | dm3.chrX |
|---|---|
| Location | 2,702,228 – 2,702,345 |
| Length | 117 |
| Max. P | 0.916207 |

| Location | 2,702,228 – 2,702,345 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 70.23 |
| Shannon entropy | 0.49551 |
| G+C content | 0.51884 |
| Mean single sequence MFE | -25.70 |
| Consensus MFE | -13.20 |
| Energy contribution | -13.20 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.916207 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
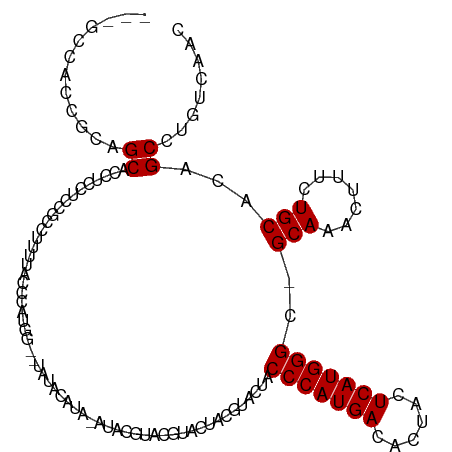
>dm3.chrX 2702228 117 + 22422827 GCCUCCUUCGCAGCUUCUCCACCGCCUUUUGCCUAUGGUACAUACAUACAUACGUACGUACUACGUACUACCCAUGACACUACUCAUGGGC-GCAAACUUUGUGCACAGCCUGUCAAC .........((((((....(((.....(((((...((((((.(((........))).)))))).......(((((((......))))))).-)))))....)))...)).)))).... ( -26.80, z-score = -1.81, R) >droSec1.super_10 2461625 96 + 3036183 ---GCCUCCGCAGCUUCUCCUCCG------------------UACAUAUAUACGUACGUACUACGUACUACCCAUGACACUACUCAUGGGC-GCAAACUUUCUGCACAGCCUGUCAAC ---......((((...........------------------...........(((((.....)))))..(((((((......))))))).-.........))))............. ( -21.40, z-score = -2.43, R) >droYak2.chrX 16127478 96 - 21770863 ---GCCACAGGAGCACUUCCUUCGCCUUUUACCCACGG--UAUGUA-----------------CGUACUACCCAUGACACUACUCAUGGGCUGCAAACUUUCUGCACAGCCUGUCAAC ---...(((((.(((........((...........((--(((...-----------------.))))).(((((((......)))))))..))........)))....))))).... ( -21.89, z-score = -1.20, R) >droEre2.scaffold_4690 136682 113 + 18748788 ---GCCACCGGAGCACCUCCCUCGCCUUUCACCCAUGGG-UAUGUAGUACGUACGUAUAGCUACGUACUACCCAUGACACUACUCAUGGGC-GCAAACUUUCUGCACAGCCUGUCAAC ---.....(((.((.................((((((((-(((((.(((.(((((((....)))))))))).....))).)))))))))).-(((.......)))...)))))..... ( -32.70, z-score = -2.01, R) >consensus ___GCCACCGCAGCACCUCCUCCGCCUUUUACCCAUGG__UAUACAUA_AUACGUACGUACUACGUACUACCCAUGACACUACUCAUGGGC_GCAAACUUUCUGCACAGCCUGUCAAC ............((........................................................(((((((......)))))))..(((.......)))...))........ (-13.20 = -13.20 + 0.00)

| Location | 2,702,228 – 2,702,345 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 70.23 |
| Shannon entropy | 0.49551 |
| G+C content | 0.51884 |
| Mean single sequence MFE | -35.17 |
| Consensus MFE | -16.38 |
| Energy contribution | -16.38 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.632505 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
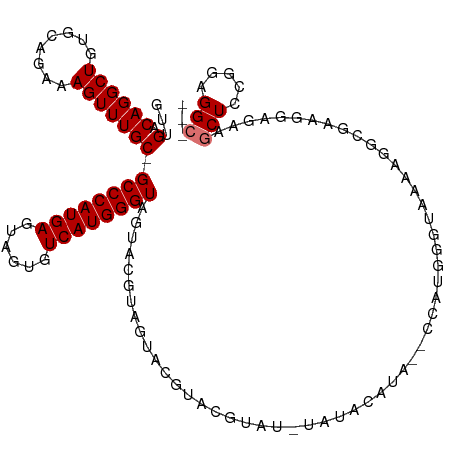
>dm3.chrX 2702228 117 - 22422827 GUUGACAGGCUGUGCACAAAGUUUGC-GCCCAUGAGUAGUGUCAUGGGUAGUACGUAGUACGUACGUAUGUAUGUAUGUACCAUAGGCAAAAGGCGGUGGAGAAGCUGCGAAGGAGGC .(((...((((.(.(((....(((((-((((((((......))))))))........((((((((((....)))))))))).....))))).....))).)..)))).)))....... ( -38.40, z-score = -1.65, R) >droSec1.super_10 2461625 96 - 3036183 GUUGACAGGCUGUGCAGAAAGUUUGC-GCCCAUGAGUAGUGUCAUGGGUAGUACGUAGUACGUACGUAUAUAUGUA------------------CGGAGGAGAAGCUGCGGAGGC--- .......(.((.(((((....(((.(-((((((((......)))))))).(((((((((((....)))).))))))------------------)...).)))..))))).)).)--- ( -34.40, z-score = -3.09, R) >droYak2.chrX 16127478 96 + 21770863 GUUGACAGGCUGUGCAGAAAGUUUGCAGCCCAUGAGUAGUGUCAUGGGUAGUACG-----------------UACAUA--CCGUGGGUAAAAGGCGAAGGAAGUGCUCCUGUGGC--- ....(((((..(..(......(((((.((((((((......))))))))..(((.-----------------(((...--..))).)))....)))))....)..).)))))...--- ( -29.80, z-score = -0.94, R) >droEre2.scaffold_4690 136682 113 - 18748788 GUUGACAGGCUGUGCAGAAAGUUUGC-GCCCAUGAGUAGUGUCAUGGGUAGUACGUAGCUAUACGUACGUACUACAUA-CCCAUGGGUGAAAGGCGAGGGAGGUGCUCCGGUGGC--- .....((((((........))))))(-(((((((.(((.(((....(((((((((((....))))))).)))))))))-).))))))))...(.((..((((...))))..)).)--- ( -38.10, z-score = -0.54, R) >consensus GUUGACAGGCUGUGCAGAAAGUUUGC_GCCCAUGAGUAGUGUCAUGGGUAGUACGUAGUACGUACGUAU_UAUACAUA__CCAUGGGUAAAAGGCGAAGGAGAAGCUCCGGAGGC___ .....((((((........))))))..((((((((......))))))))..................................................................... (-16.38 = -16.38 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:10:09 2011