| Sequence ID | dm3.chrX |
|---|---|
| Location | 2,665,543 – 2,665,642 |
| Length | 99 |
| Max. P | 0.999208 |

| Location | 2,665,543 – 2,665,642 |
|---|---|
| Length | 99 |
| Sequences | 11 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 76.79 |
| Shannon entropy | 0.44968 |
| G+C content | 0.45218 |
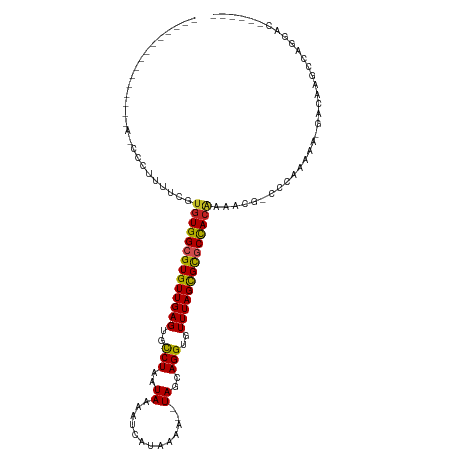
| Mean single sequence MFE | -29.40 |
| Consensus MFE | -24.60 |
| Energy contribution | -24.50 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.29 |
| Mean z-score | -3.25 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.71 |
| SVM RNA-class probability | 0.999208 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
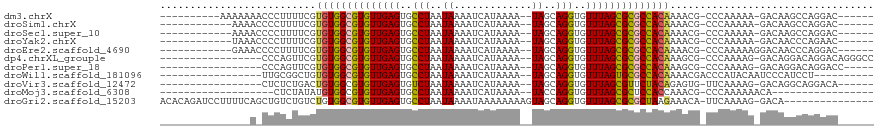
>dm3.chrX 2665543 99 - 22422827 ----------AAAAAAACCCUUUUCGUGUGGCGUGUUGAGUGCCUAAUAAAAUCAUAAAA--UAGCAGGUGUUUAGCGCGCCACAAAACG-CCCAAAAA-GACAAGCCAGGAC------ ----------........(((.....((((((((((((((..(((..((...........--))..)))..))))))))))))))....(-(.......-.....)).)))..------ ( -29.20, z-score = -3.28, R) >droSim1.chrX 1852220 97 - 17042790 ------------AAAACCCCUUUUCGUGUGGCGUGUUGAGUGCCUAAUAAAAUCAUAAAA--UAGCAGGUGUUUAGCGCGCCACAAAACG-CCCAAAAA-GACAAGCCAGGAC------ ------------....((.(((.((.((((((((((((((..(((..((...........--))..)))..)))))))))))))).....-........-)).)))...))..------ ( -29.34, z-score = -3.34, R) >droSec1.super_10 2425761 97 - 3036183 ------------AAAACCCCUUUUCGUGUGGCGUGUUGAGUGCCUAAUAAAAUCAUAAAA--UAGCAGGUGUUUAGCGCGCCACAAAACG-CCCAAAAA-GACAAGCCAGGAC------ ------------....((.(((.((.((((((((((((((..(((..((...........--))..)))..)))))))))))))).....-........-)).)))...))..------ ( -29.34, z-score = -3.34, R) >droYak2.chrX 16088375 97 + 21770863 ------------UAAACCCCUUUUCGUGUGGCGUGUUGAGUGCCUAAUAAAAUCAUAAAA--UAGCAGGUGUUUAGCGCGCCACAAAACG-CCCAAAAA-GACAACCCAGAAC------ ------------.......(((((((((((((((((((((..(((..((...........--))..)))..)))))))))))))...)))-....))))-)............------ ( -28.70, z-score = -4.20, R) >droEre2.scaffold_4690 92339 98 - 18748788 ------------GAAACCCCUUUUCGUGUGGCGUGUUGAGUGCCUAAUAAAAUCAUAAAA--UAGCAGGUGUUUAGCGCGCCACAAAACG-CCCAAAAAGGACAACCCAGGAC------ ------------......((((((((((((((((((((((..(((..((...........--))..)))..)))))))))))))...)))-....))))))............------ ( -32.00, z-score = -3.85, R) >dp4.chrXL_group1e 2049639 98 - 12523060 -----------------CCCAGUUCGUGUGGCGUGUUGAGUGCCUAAUAAAAUCAUAAAA--UAGCAGGUGUUUAGCGCGCCACAAAGCG-CCCAAAAG-GACAGGACAGGACAGGGCC -----------------(((.((((.((((((((((((((..(((..((...........--))..)))..))))))))))))))....(-.((....)-).)......)))).))).. ( -37.30, z-score = -3.78, R) >droPer1.super_18 903674 93 - 1952607 -----------------CCCAGUUCGUGUGGCGUGUUGAGUGCCUAAUAAAAUCAUAAAA--UAGCAGGUGUUUAGCGCGCCACAAAGCG-CCCAAAAG-GACAGGACAGGACC----- -----------------.((.((((.((((((((((((((..(((..((...........--))..)))..))))))))))))))....(-.((....)-).).)))).))...----- ( -35.00, z-score = -4.27, R) >droWil1.scaffold_181096 1998175 90 - 12416693 -----------------UUGCGGCUGUGUGGCGUGUUGAGUGCCUAAUAAAAUCAUAAAA--UAGCAGGUGUUUAGUGCGCCACAAAACGACCCAUACAAUCCCAUCCU---------- -----------------...((....(((((((..(((((..(((..((...........--))..)))..)))))..)))))))...))...................---------- ( -27.40, z-score = -2.53, R) >droVir3.scaffold_12472 694064 92 + 763072 -----------------CUCUCUGACUGUGGCGUGUUGAGUGUCUAAUAAAAUCAUAAAA--UAGCAGGUGUUUAGCGUUCUACAGAGUG-UUCAAAAG-GACAGGCAGGACA------ -----------------.((.(((.((((((..(((((((..(((..((...........--))..)))..)))))))..))))))..((-(((....)-))))..)))))..------ ( -23.60, z-score = -1.04, R) >droMoj3.scaffold_6308 1506505 80 - 3356042 -------------------CUCUAUAUGUGGCGUGUUGAGUGCCUAAUAAAAUCAUAAAA--UACCAGGUGUUUAGCGCUCCACCAAACG-CCCAAAAAACA----------------- -------------------........((((.((((((((..(((...............--....)))..)))))))).))))......-...........----------------- ( -20.31, z-score = -3.07, R) >droGri2.scaffold_15203 2140911 102 - 11997470 ACACAGAUCCUUUUCAGCUGUCUGUCUGUGGCGUGUUGAGUGCCUAAUAAAAUAAAAAAAAGUAGCAGGUGUUUAGCGCGCUAAGAAACA-UUCAAAAG-GACA--------------- .......(((((((.((.(((...(((.((((((((((((..(((..((.............))..)))..))))))))))))))).)))-)).)))))-))..--------------- ( -31.22, z-score = -3.03, R) >consensus _______________A_CCCUUUUCGUGUGGCGUGUUGAGUGCCUAAUAAAAUCAUAAAA__UAGCAGGUGUUUAGCGCGCCACAAAACG_CCCAAAAA_GACAAGCCAGGAC______ ..........................((((((((((((((..(((.....................)))..)))))))))))))).................................. (-24.60 = -24.50 + -0.10)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:10:05 2011