
| Sequence ID | dm3.chrX |
|---|---|
| Location | 2,642,350 – 2,642,470 |
| Length | 120 |
| Max. P | 0.856163 |

| Location | 2,642,350 – 2,642,470 |
|---|---|
| Length | 120 |
| Sequences | 7 |
| Columns | 128 |
| Reading direction | forward |
| Mean pairwise identity | 77.05 |
| Shannon entropy | 0.44868 |
| G+C content | 0.42673 |
| Mean single sequence MFE | -33.84 |
| Consensus MFE | -12.01 |
| Energy contribution | -14.38 |
| Covariance contribution | 2.37 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.856163 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 2642350 120 + 22422827 ------GCCCUUAUUCACGGUAUCCUCGAGG--CAAGUUGAUUGUAUAAAGCAAAACUAUAGCCGAGGAUACAGCAAAGUAAACAACUUACUUUGUAAUCAGAGCCCAGGAUUGAGUGCUGCAAGAAC ------((.((((.((...((((((((..((--(.((((..((((.....))))))))...))))))))))).(((((((((.....))))))))).............)).)))).))......... ( -33.90, z-score = -2.94, R) >droSim1.chrX 1831751 120 + 17042790 ------GGCCUUAUUCACGGUAUCCUCGAGG--CAAGUUGAUUGUAUAAAGCAAAACUAUAGCCGAGGAUACAGCAAAGUAAACAACUUACUUUGUAAUCAGAGCCCGGGAUUGGGUGCUGCAAGAAC ------.((((.((((.((((((((((..((--(.((((..((((.....))))))))...))))))))))).(((((((((.....)))))))))..........)))))).))))........... ( -36.70, z-score = -2.80, R) >droSec1.super_10 2403475 120 + 3036183 ------GGCCUUAUUCACGGUAUCCUCGAGG--CAAGUUGAUUGUAUAAAGGAAAACUAUAGCCGAGGAUACAGCAAAGUAAACAACUUACUUUGUAAUCAGAGCCCAGGAUUGGGUGCUGCAAGAAC ------.......(((...((((((((..((--(.((((..((......))...))))...))))))))))).(((((((((.....)))))))))...(((.((((......)))).)))...))). ( -35.20, z-score = -2.57, R) >droYak2.chrX 16063016 120 - 21770863 ------GGCUAUAUUCACGGUAUCCUCGAGC--CAAGUUGAUUGUAUAAAGCAAAACUAUAGCCGAGGAUACAGCAAAGUAAACAACUUACUUUGUAAUCAGAGCCCAGGAUUGGGUGCUGCAAGAAC ------.......(((...(((((((((.((--..((((..((((.....))))))))...))))))))))).(((((((((.....)))))))))...(((.((((......)))).)))...))). ( -38.50, z-score = -3.98, R) >droEre2.scaffold_4690 68817 120 + 18748788 ------GGCUAUAUUCACGGUAUCCUCGAGC--CAAGUUGAUUGUAUAAAGCAAAACUAUAGCCGAGGAUACAGCAAAGUAAACAACUUACUUUGUAAUCAGAGCCCAGGAUUGGGUGCUGCAGGAAC ------.......(((...(((((((((.((--..((((..((((.....))))))))...))))))))))).(((((((((.....)))))))))...(((.((((......)))).)))..))).. ( -38.30, z-score = -3.54, R) >droAna3.scaffold_13248 4412261 117 - 4840945 AAAUAAUAGCAUAUUCAUGGUGUCUUUGGACACCGAGUUGAUUGUAUAAAG-----------CCCAGACUGGAGCAAAGUAAACGGCUUACUUUGUAAUCAGAGCCAAGGAUUUUGAGAUGCAACAAC ........((((.((((.((((((....))))))(..((((((........-----------.((.....)).(((((((((.....)))))))))))))))..).........))))))))...... ( -30.60, z-score = -2.19, R) >droWil1.scaffold_181096 1960216 101 + 12416693 -------------GUCCUGCU-UCAAGGCUCGCCAAGUUGAUUGUAUAUAUAAAG--------CAAAACUACACUC---UUAACGGCUUACUUUGUAAUCAGAGCCAAGAAUCCUGCA--GCGAGGAC -------------((((((((-.(..((((((((.....((.((((.........--------......)))).))---.....)))((((...))))...))))).((....))).)--)).))))) ( -23.66, z-score = -0.97, R) >consensus ______GGCCAUAUUCACGGUAUCCUCGAGC__CAAGUUGAUUGUAUAAAGCAAAACUAUAGCCGAGGAUACAGCAAAGUAAACAACUUACUUUGUAAUCAGAGCCCAGGAUUGGGUGCUGCAAGAAC ...................(((((((((...................................))))))))).(((((((((.....)))))))))......(((((.......)).)))........ (-12.01 = -14.38 + 2.37)

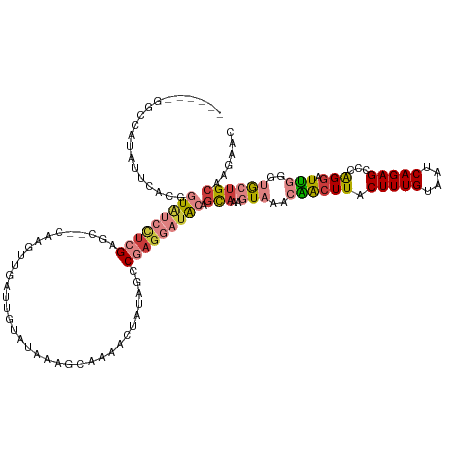
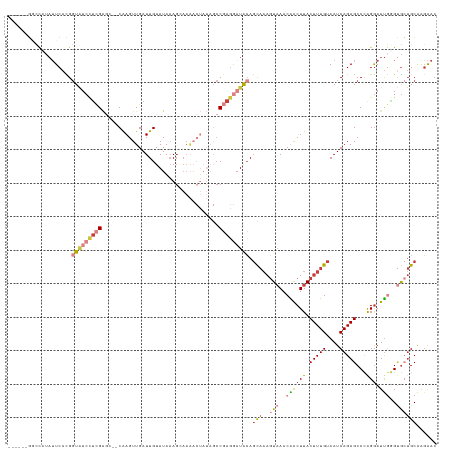
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:10:01 2011