
| Sequence ID | dm3.chrX |
|---|---|
| Location | 2,623,817 – 2,623,933 |
| Length | 116 |
| Max. P | 0.731978 |

| Location | 2,623,817 – 2,623,933 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 57.70 |
| Shannon entropy | 0.55414 |
| G+C content | 0.61381 |
| Mean single sequence MFE | -40.07 |
| Consensus MFE | -26.05 |
| Energy contribution | -25.07 |
| Covariance contribution | -0.98 |
| Combinations/Pair | 1.44 |
| Mean z-score | -0.69 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.731978 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
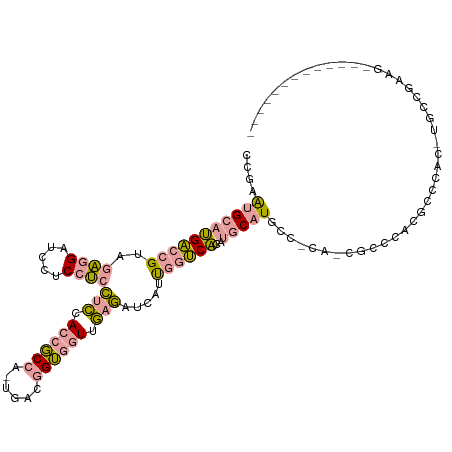
>dm3.chrX 2623817 116 + 22422827 CAUCAAUCCUCCCAGCCGACAAGCGGGCGCAGAAGGUGCGACAGGCGGAGGUGCGACAGGUGGAAGUCCGACCGGUGGAGGUGCGACCGGUGGAUCGGCAACCAACCUUCACAUAA .......((((((..(((.....)))((((.....)))).....).)))))((.((.((((((..((((.((((((.(.....).)))))).)...)))..)).)))))).))... ( -39.00, z-score = 0.83, R) >dp4.chrXL_group1e 2015352 99 + 12523060 -------------CUUCGGCA-GUGGGCGUGGGCG-UG-GGCAUGCAUGUGACCAAUGAUCUCAACCACCGUCA-UGGCGGUGGAGGAGGAGGAUCCUCUACGGUCAUGCAUUCGG -------------...(((((-((..(((....))-).-.)).)))..((((((......(((..(((((((..-..)))))))..)))(((....)))...)))))).....)). ( -38.60, z-score = -0.84, R) >droPer1.super_18 865939 99 + 1952607 -------------CUUCGGCA-GUCGGCGUGGGCG-UG-GGCAUGCAUGUGACCAAUGAUCUCAACCACCGUCA-UGGCGGUGGAGGAGGAGGAUCCUCUACGGUCAUGCAUUCGG -------------...(((((-(((.(((....))-).-))).)))..((((((......(((..(((((((..-..)))))))..)))(((....)))...)))))).....)). ( -42.60, z-score = -2.06, R) >consensus _____________CUUCGGCA_GUGGGCGUGGGCG_UG_GGCAUGCAUGUGACCAAUGAUCUCAACCACCGUCA_UGGCGGUGGAGGAGGAGGAUCCUCUACGGUCAUGCAUUCGG ..........................(((((..........))))).(((((((......(((..(((((((.....)))))))..)))(((....)))...)))))))....... (-26.05 = -25.07 + -0.98)



| Location | 2,623,817 – 2,623,933 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 57.70 |
| Shannon entropy | 0.55414 |
| G+C content | 0.61381 |
| Mean single sequence MFE | -34.03 |
| Consensus MFE | -16.67 |
| Energy contribution | -17.23 |
| Covariance contribution | 0.57 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.04 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chrX 2623817 116 - 22422827 UUAUGUGAAGGUUGGUUGCCGAUCCACCGGUCGCACCUCCACCGGUCGGACUUCCACCUGUCGCACCUCCGCCUGUCGCACCUUCUGCGCCCGCUUGUCGGCUGGGAGGAUUGAUG ...(((((((((.((...((((....(((((.(.....).)))))))))....)))))).)))))(((((.......(((.....)))(((........)))..)))))....... ( -38.50, z-score = 0.28, R) >dp4.chrXL_group1e 2015352 99 - 12523060 CCGAAUGCAUGACCGUAGAGGAUCCUCCUCCUCCACCGCCA-UGACGGUGGUUGAGAUCAUUGGUCACAUGCAUGCC-CA-CGCCCACGCCCAC-UGCCGAAG------------- .((.(((((((((((..((((.....))))(((.((((((.-....)))))).))).....))))))..)))))((.-..-.((....))....-.))))...------------- ( -31.80, z-score = -1.80, R) >droPer1.super_18 865939 99 - 1952607 CCGAAUGCAUGACCGUAGAGGAUCCUCCUCCUCCACCGCCA-UGACGGUGGUUGAGAUCAUUGGUCACAUGCAUGCC-CA-CGCCCACGCCGAC-UGCCGAAG------------- .((.(((((((((((..((((.....))))(((.((((((.-....)))))).))).....))))))..)))))((.-..-.((....))....-.))))...------------- ( -31.80, z-score = -1.60, R) >consensus CCGAAUGCAUGACCGUAGAGGAUCCUCCUCCUCCACCGCCA_UGACGGUGGUUGAGAUCAUUGGUCACAUGCAUGCC_CA_CGCCCACGCCCAC_UGCCGAAG_____________ ....(((((((((((..((((.....))))(((.((((((......)))))).))).....))))))..))))).......................................... (-16.67 = -17.23 + 0.57)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:09:59 2011