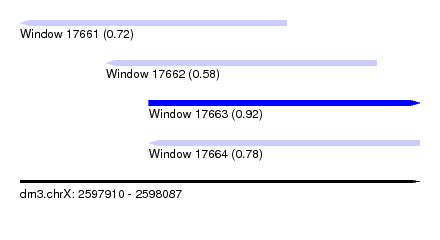
| Sequence ID | dm3.chrX |
|---|---|
| Location | 2,597,910 – 2,598,087 |
| Length | 177 |
| Max. P | 0.917607 |

| Location | 2,597,910 – 2,598,028 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.67 |
| Shannon entropy | 0.42983 |
| G+C content | 0.41273 |
| Mean single sequence MFE | -34.72 |
| Consensus MFE | -14.89 |
| Energy contribution | -15.17 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.42 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.716151 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 2597910 118 - 22422827 AAUAGCAACAAAAGCUGCACGCCAUGGGCGUAGCUUGCUUUAAAAUGGGGAUCCAUCCCCCAGGAGCAUAUGUAAGGUGUAUUUA--CUUUACUGAAAUGCACAAUGCACUUUUAAAUUU ....(((...(((((.(((((((...))))).))..)))))....(((((.......)))))...((((..((((((((....))--))))))....))))....)))............ ( -36.40, z-score = -2.47, R) >droEre2.scaffold_4690 24446 116 - 18748788 AGCAGCAACAAAAGUUGCGGGCCAUGGGCGU-GCAUGGUAUAGAAAGGGGAUUU-UCCCCCAAGCGCAUAUUUAAGGUGUAGGUA--UUUUAGUGAAACACUCCAUGCGCCUUUGAACUU .((.(((((....)))))..))((.((((..-((((((........((((....-..))))..............(((((...((--(....)))..))))))))))))))).))..... ( -35.60, z-score = -0.93, R) >droYak2.chrX 16018482 114 + 21770863 AGCAGCAACAAAAGUUGCAGGCCAUGGGCGUAGCCUGGUAUAGAAAAGGGAUAUUUCGCCCGAGAACAUAUUCAAGGAGUAUUUAUGUUUAAGUGAAAUAUGGAGUAUUCAUUU------ ....(((((....))))).(..(.((((((...(((..(.....)..)))......)))))).)..).........((((((((((((((.....)))))).))))))))....------ ( -27.70, z-score = -1.49, R) >droSec1.super_10 2359485 118 - 3036183 AACAGCAACAAAAGUUGCAGGACAUGGGCGUAGUUUGCUAUAUGAUGGGGGUCCAUCCCCCAGAAGCAUAUGUAAGCUAUAUUUA--CUUUACUGAAAUUCACAAAGCACUUUUAAAUUU ....(((((....))))).........(((((((((((.(((((.((((((.....))))))....)))))))))))))).....--......((.....))....))............ ( -33.60, z-score = -2.70, R) >droSim1.chrX 1801030 118 - 17042790 AACAGCAACAAAAGUUGCAGGCCAUGGGCGUAGUUUGCUAUAUGAUGGGGGUCCAUCCCCCAGAAGCAUAUGUAAGCUAUAUUUA--CUUUACUGAAAUGCACAAUGCAGCUCUAAAUUU ....(((((....))))).......(((((((((((((.(((((.((((((.....))))))....)))))))))))))).....--...........(((.....)))))))....... ( -40.30, z-score = -3.23, R) >consensus AACAGCAACAAAAGUUGCAGGCCAUGGGCGUAGCUUGCUAUAGAAUGGGGAUCCAUCCCCCAGGAGCAUAUGUAAGGUGUAUUUA__CUUUACUGAAAUACACAAUGCACCUUUAAAUUU .((.(((((....)))))..(((...)))))...............((((.......))))............((((((((((....................))))))))))....... (-14.89 = -15.17 + 0.28)

| Location | 2,597,948 – 2,598,068 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.50 |
| Shannon entropy | 0.30401 |
| G+C content | 0.49336 |
| Mean single sequence MFE | -40.92 |
| Consensus MFE | -23.57 |
| Energy contribution | -24.73 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.584584 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 2597948 120 - 22422827 CCAGCACUGUGACUAUCGAAAAGUCGGUCUCUGCUGCGAAAAUAGCAACAAAAGCUGCACGCCAUGGGCGUAGCUUGCUUUAAAAUGGGGAUCCAUCCCCCAGGAGCAUAUGUAAGGUGU ...(((((.(((((.......)))))((..(((.(((.......)))...(((((.(((((((...))))).))..))))).....(((((....))))))))..))........))))) ( -36.90, z-score = -0.91, R) >droEre2.scaffold_4690 24484 118 - 18748788 GCAUCACUGUGGCUAUCGAAAUGUCGGCUUCUGCCGCAAAAGCAGCAACAAAAGUUGCGGGCCAUGGGCGU-GCAUGGUAUAGAAAGGGGAU-UUUCCCCCAAGCGCAUAUUUAAGGUGU (((((.((((((((.......((.((((....))))))......(((((....))))).))))))))..((-((.(((.........(((..-...)))))).))))........))))) ( -42.30, z-score = -1.60, R) >droYak2.chrX 16018516 120 + 21770863 GCAGCACUGUGGCUAUCGAAAAGUCGGCCUCUGCCACAAAAGCAGCAACAAAAGUUGCAGGCCAUGGGCGUAGCCUGGUAUAGAAAAGGGAUAUUUCGCCCGAGAACAUAUUCAAGGAGU ...((.((((((((........((.(((....))))).......(((((....))))).))))))))))....(((...(((.....(((........))).......)))...)))... ( -33.10, z-score = -0.05, R) >droSec1.super_10 2359523 120 - 3036183 UCAGCACUGUGGCUAUCGAAAAGUCGGUCUCUGCUGCGAAAACAGCAACAAAAGUUGCAGGACAUGGGCGUAGUUUGCUAUAUGAUGGGGGUCCAUCCCCCAGAAGCAUAUGUAAGCUAU ((((..(((..((((((((....)))))...(((((......))))).....)))..)))..).)))..(((((((((.(((((.((((((.....))))))....)))))))))))))) ( -45.00, z-score = -3.36, R) >droSim1.chrX 1801068 120 - 17042790 UCAGCACUGUGGUUAUCGAAAAGUCUGUCUCUGCUGCGAAAACAGCAACAAAAGUUGCAGGCCAUGGGCGUAGUUUGCUAUAUGAUGGGGGUCCAUCCCCCAGAAGCAUAUGUAAGCUAU ...((.((((((((.(((...(((........))).))).....(((((....))))).))))))))))(((((((((.(((((.((((((.....))))))....)))))))))))))) ( -47.30, z-score = -4.17, R) >consensus GCAGCACUGUGGCUAUCGAAAAGUCGGUCUCUGCUGCGAAAACAGCAACAAAAGUUGCAGGCCAUGGGCGUAGCUUGCUAUAGAAUGGGGAUCCAUCCCCCAGGAGCAUAUGUAAGGUGU ...((.((((((((........(.((((....))))).......(((((....))))).))))))))))....(((((.......(((((.......))))).........))))).... (-23.57 = -24.73 + 1.16)


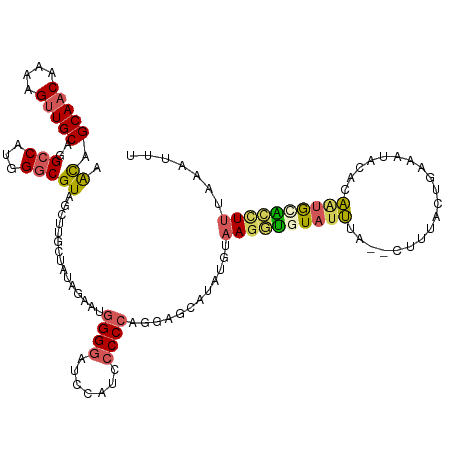
| Location | 2,597,967 – 2,598,087 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.33 |
| Shannon entropy | 0.27559 |
| G+C content | 0.54186 |
| Mean single sequence MFE | -44.00 |
| Consensus MFE | -31.00 |
| Energy contribution | -31.12 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.917607 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 2597967 120 + 22422827 GGGGGAUGGAUCCCCAUUUUAAAGCAAGCUACGCCCAUGGCGUGCAGCUUUUGUUGCUAUUUUCGCAGCAGAGACCGACUUUUCGAUAGUCACAGUGCUGGCAUUGUGGCGGAACUCGCG .(((((....)))))........((((((((((((...)))))..))))).((((((.......))))))(((..(((....)))...(((((((((....)))))))))....))))). ( -45.30, z-score = -2.29, R) >droEre2.scaffold_4690 24503 118 + 18748788 -GGGGGAAAAUCCCCUUUCUAUACCAUGC-ACGCCCAUGGCCCGCAACUUUUGUUGCUGCUUUUGCGGCAGAAGCCGACAUUUCGAUAGCCACAGUGAUGCCAUUGUGGCGGGAGUCGCG -(((((.....)))))...........((-((.(((..(((..(((((....))))).)))..((((((....)))).))........(((((((((....)))))))))))).)).)). ( -47.50, z-score = -2.80, R) >droYak2.chrX 16018535 120 - 21770863 GGGCGAAAUAUCCCUUUUCUAUACCAGGCUACGCCCAUGGCCUGCAACUUUUGUUGCUGCUUUUGUGGCAGAGGCCGACUUUUCGAUAGCCACAGUGCUGCCAUUGCAGCGGGACUUGCG ..((((....((((............(((((((....(((((((((((....)))))((((.....)))).))))))......)).))))).....(((((....))))))))).)))). ( -44.40, z-score = -2.07, R) >droSec1.super_10 2359542 120 + 3036183 GGGGGAUGGACCCCCAUCAUAUAGCAAACUACGCCCAUGUCCUGCAACUUUUGUUGCUGUUUUCGCAGCAGAGACCGACUUUUCGAUAGCCACAGUGCUGACAUUGUGGCGGGACUCGCG (((((.....)))))................(((....(((((.....(((((((((.......)))))))))..(((....)))...(((((((((....))))))))))))))..))) ( -44.40, z-score = -2.87, R) >droSim1.chrX 1801087 120 + 17042790 GGGGGAUGGACCCCCAUCAUAUAGCAAACUACGCCCAUGGCCUGCAACUUUUGUUGCUGUUUUCGCAGCAGAGACAGACUUUUCGAUAACCACAGUGCUGACAUUGUGGCGGGACUCGCG (((((.....)))))................(((....(((((((...(((((((((.......))))))))).................(((((((....)))))))))))).)).))) ( -38.40, z-score = -1.31, R) >consensus GGGGGAUGGAUCCCCAUUAUAUAGCAAGCUACGCCCAUGGCCUGCAACUUUUGUUGCUGUUUUCGCAGCAGAGACCGACUUUUCGAUAGCCACAGUGCUGACAUUGUGGCGGGACUCGCG (((((.....)))))................(((......(((((...(((((((((.......))))))))).................(((((((....))))))))))))....))) (-31.00 = -31.12 + 0.12)

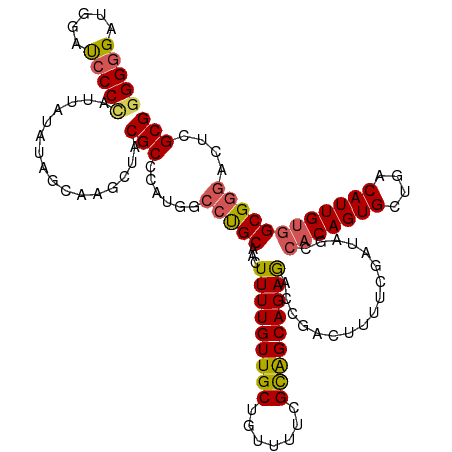
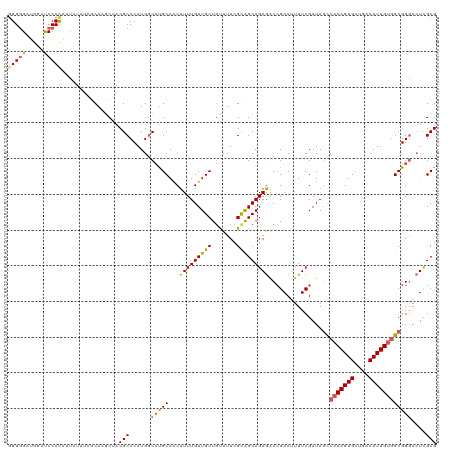
| Location | 2,597,967 – 2,598,087 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.33 |
| Shannon entropy | 0.27559 |
| G+C content | 0.54186 |
| Mean single sequence MFE | -42.22 |
| Consensus MFE | -32.64 |
| Energy contribution | -34.04 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.784800 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 2597967 120 - 22422827 CGCGAGUUCCGCCACAAUGCCAGCACUGUGACUAUCGAAAAGUCGGUCUCUGCUGCGAAAAUAGCAACAAAAGCUGCACGCCAUGGGCGUAGCUUGCUUUAAAAUGGGGAUCCAUCCCCC .(((((((.((((.((..((((((...(.((((..(.....)..)))).)(((((......)))))......))))...))..)))))).)))))))........(((((....))))). ( -40.00, z-score = -2.04, R) >droEre2.scaffold_4690 24503 118 - 18748788 CGCGACUCCCGCCACAAUGGCAUCACUGUGGCUAUCGAAAUGUCGGCUUCUGCCGCAAAAGCAGCAACAAAAGUUGCGGGCCAUGGGCGU-GCAUGGUAUAGAAAGGGGAUUUUCCCCC- .((.((.((((((.....)))......((((((.......((.((((....))))))......(((((....))))).))))))))).))-))............((((.....)))).- ( -44.80, z-score = -1.71, R) >droYak2.chrX 16018535 120 + 21770863 CGCAAGUCCCGCUGCAAUGGCAGCACUGUGGCUAUCGAAAAGUCGGCCUCUGCCACAAAAGCAGCAACAAAAGUUGCAGGCCAUGGGCGUAGCCUGGUAUAGAAAAGGGAUAUUUCGCCC .....((((((((((....)))))((((.(((((.((....((.(((((.(((.......)))(((((....))))))))))))...)))))))))))........)))))......... ( -44.00, z-score = -1.69, R) >droSec1.super_10 2359542 120 - 3036183 CGCGAGUCCCGCCACAAUGUCAGCACUGUGGCUAUCGAAAAGUCGGUCUCUGCUGCGAAAACAGCAACAAAAGUUGCAGGACAUGGGCGUAGUUUGCUAUAUGAUGGGGGUCCAUCCCCC .((((((..((((...(((((....(((..((((((((....)))))...(((((......))))).....)))..)))))))).))))..))))))........(((((.....))))) ( -44.30, z-score = -2.24, R) >droSim1.chrX 1801087 120 - 17042790 CGCGAGUCCCGCCACAAUGUCAGCACUGUGGUUAUCGAAAAGUCUGUCUCUGCUGCGAAAACAGCAACAAAAGUUGCAGGCCAUGGGCGUAGUUUGCUAUAUGAUGGGGGUCCAUCCCCC .(((.....)))......((((((.((((((((.(((...(((........))).))).....(((((....))))).))))))))))((((....)))).))))(((((.....))))) ( -38.00, z-score = -0.78, R) >consensus CGCGAGUCCCGCCACAAUGGCAGCACUGUGGCUAUCGAAAAGUCGGUCUCUGCUGCGAAAACAGCAACAAAAGUUGCAGGCCAUGGGCGUAGCUUGCUAUAGAAUGGGGAUCCAUCCCCC .((((((..((((...(((((.....((((((.(((((....)))))....))))))......(((((....)))))..))))).))))..))))))........(((((....))))). (-32.64 = -34.04 + 1.40)

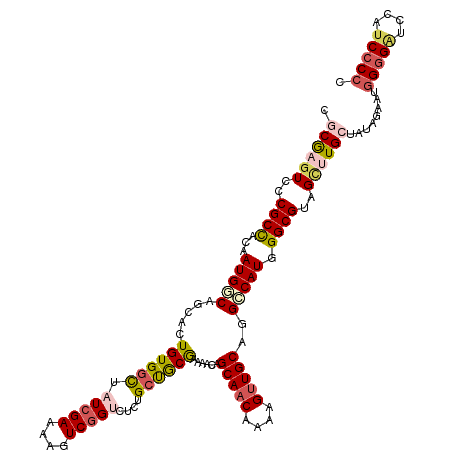
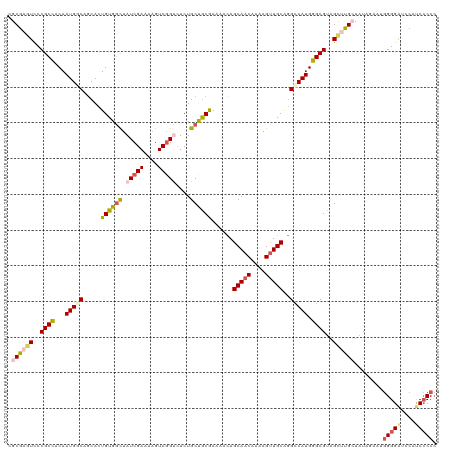
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:09:56 2011