
| Sequence ID | dm3.chrX |
|---|---|
| Location | 2,597,137 – 2,597,233 |
| Length | 96 |
| Max. P | 0.538821 |

| Location | 2,597,137 – 2,597,233 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 65.87 |
| Shannon entropy | 0.57920 |
| G+C content | 0.52652 |
| Mean single sequence MFE | -24.83 |
| Consensus MFE | -11.02 |
| Energy contribution | -11.25 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.517518 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
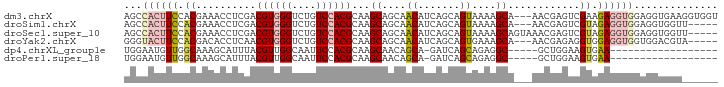
>dm3.chrX 2597137 96 + 22422827 ACCACCUUCACCUCCACCUCUUCGACUCGUU---UGCUUUUACUGCUGAUGUUGCUGCUUGCGUGGACAGACCCACGUCGAGGUUUCGUGGAAGUGGCU .......((((.(((((..(((((((.((((---.((.......)).))))..((.....))((((......)))))))))))....))))).)))).. ( -31.60, z-score = -2.29, R) >droSim1.chrX 1800267 91 + 17042790 -----AACCACCUCCACCUCUACGACUCGUU---UGCUUUUACUGCUGAUGUUGCUGCUUGCGUGGACAGACCCACGUCGAGGUUUCGUGGAAGUGGCU -----........((((.(((((((..((((---.((.......)).))))......(((((((((......))))).))))...))))))).)))).. ( -30.30, z-score = -2.10, R) >droSec1.super_10 2358721 94 + 3036183 -----AACCACCUCCACCUCUACGACUCGUUUACUGCUUUUACUGCUGAUGUUGCUGCUUGCGUGGACAGACCCACGUCGAGGUUUCGUGGAAGUGGCU -----........((((.(((((((..(((..((.((.......))....)).))..(((((((((......))))).)))))..))))))).)))).. ( -28.90, z-score = -1.65, R) >droYak2.chrX 16017681 91 - 21770863 -----UACGUCCACCACCUCCACCUCUCGUU---UGCUUUCACUGCUGAUGUUGCUGCUUGCGUGGACAGACCCACGUUGAGGUGUCGUGGAAGUACCC -----(((.(((((((((((((.(...((((---.((.......)).))))..).))...((((((......)))))).))))))..))))).)))... ( -29.60, z-score = -1.76, R) >dp4.chrXL_group1e 1992664 75 + 12523060 ------------------UUCACUUCCAGC-----GCCUCUGCUGAUC-UGCUGUUGCUUGCGUGGAAUUGCCAACGUAAAUGCUUUGCCAACAUUCCA ------------------........((((-----(....)))))...-.((....((((((((((.....)).))))))..))...)).......... ( -14.30, z-score = 0.36, R) >droPer1.super_18 843040 75 + 1952607 ------------------UUCACUUCCAGC-----GCCUCUGCUGAUC-UGCUGUUGCUUGCGUGGAAUUGCCAACGUAAAUGCUUUGCCAACAUUCCA ------------------........((((-----(....)))))...-.((....((((((((((.....)).))))))..))...)).......... ( -14.30, z-score = 0.36, R) >consensus ______ACCACCUCCACCUCCACGACUCGUU___UGCUUUUACUGCUGAUGUUGCUGCUUGCGUGGACAGACCCACGUCGAGGUUUCGUGGAAGUGCCU ...............(((((...............((..(.((.......)).)..))..((((((......)))))).)))))............... (-11.02 = -11.25 + 0.23)



| Location | 2,597,137 – 2,597,233 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 65.87 |
| Shannon entropy | 0.57920 |
| G+C content | 0.52652 |
| Mean single sequence MFE | -27.62 |
| Consensus MFE | -11.10 |
| Energy contribution | -11.80 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.538821 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 2597137 96 - 22422827 AGCCACUUCCACGAAACCUCGACGUGGGUCUGUCCACGCAAGCAGCAACAUCAGCAGUAAAAGCA---AACGAGUCGAAGAGGUGGAGGUGAAGGUGGU .(((((((.(((...(((((((((((((....)))))((..((.((.......)).))....)).---.....)))...)))))....))).))))))) ( -30.20, z-score = -1.24, R) >droSim1.chrX 1800267 91 - 17042790 AGCCACUUCCACGAAACCUCGACGUGGGUCUGUCCACGCAAGCAGCAACAUCAGCAGUAAAAGCA---AACGAGUCGUAGAGGUGGAGGUGGUU----- ((((((((((((....(..(((((((((....)))))((..((.((.......)).))....)).---.....))))..)..))))))))))))----- ( -32.40, z-score = -2.66, R) >droSec1.super_10 2358721 94 - 3036183 AGCCACUUCCACGAAACCUCGACGUGGGUCUGUCCACGCAAGCAGCAACAUCAGCAGUAAAAGCAGUAAACGAGUCGUAGAGGUGGAGGUGGUU----- ((((((((((((....(..(((((((((....)))))((..((.((.......)).))....)).........))))..)..))))))))))))----- ( -32.40, z-score = -2.53, R) >droYak2.chrX 16017681 91 + 21770863 GGGUACUUCCACGACACCUCAACGUGGGUCUGUCCACGCAAGCAGCAACAUCAGCAGUGAAAGCA---AACGAGAGGUGGAGGUGGUGGACGUA----- ...(((.(((((..((((((..((((((....))))))..........((((....((.......---.))....))))))))))))))).)))----- ( -29.30, z-score = -0.61, R) >dp4.chrXL_group1e 1992664 75 - 12523060 UGGAAUGUUGGCAAAGCAUUUACGUUGGCAAUUCCACGCAAGCAACAGCA-GAUCAGCAGAGGC-----GCUGGAAGUGAA------------------ ((((((((..((...........))..)).))))))(((..((....)).-..(((((......-----)))))..)))..------------------ ( -20.70, z-score = -0.70, R) >droPer1.super_18 843040 75 - 1952607 UGGAAUGUUGGCAAAGCAUUUACGUUGGCAAUUCCACGCAAGCAACAGCA-GAUCAGCAGAGGC-----GCUGGAAGUGAA------------------ ((((((((..((...........))..)).))))))(((..((....)).-..(((((......-----)))))..)))..------------------ ( -20.70, z-score = -0.70, R) >consensus AGCCACUUCCACGAAACCUCGACGUGGGUCUGUCCACGCAAGCAGCAACAUCAGCAGUAAAAGCA___AACGAGACGUAGAGGUGGAGGUGGU______ ...((((((.((..........((((((....))))))...((....((.......))....))............)).)))))).............. (-11.10 = -11.80 + 0.70)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:09:52 2011